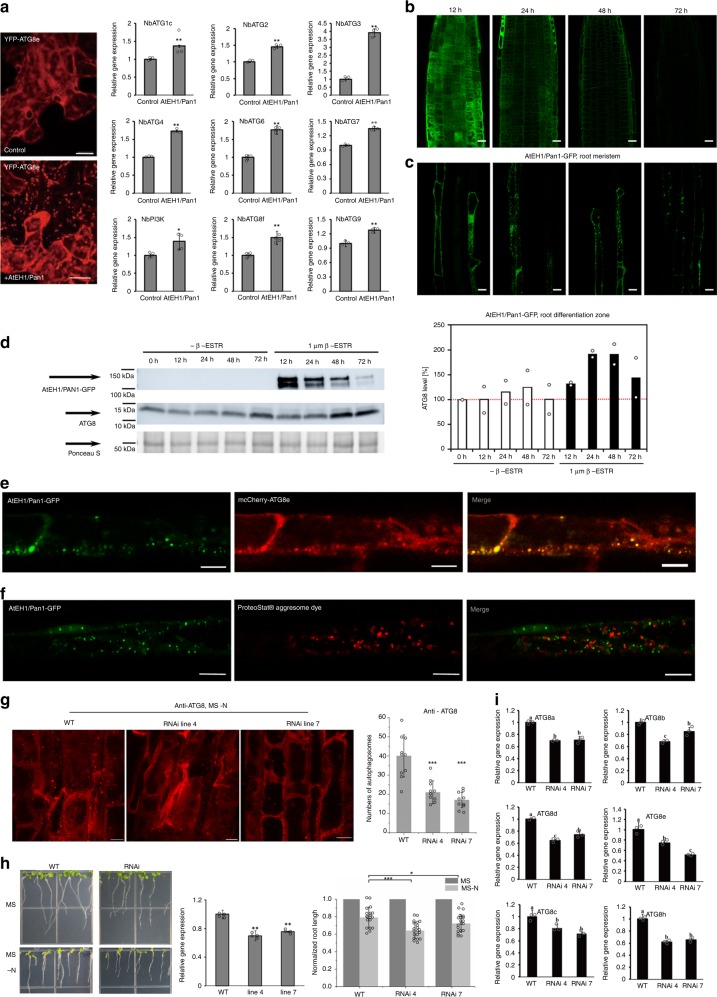
Fig. 6.
Altered expression of VAP27-1 and AtEH1/Pan1 affects autophagic activity. a RT-qPCR of selected autophagy-related genes in N. benthamiana leaves transiently expressing either GFP-AtEH1/Pan1 or empty vectors (control). In agreement with the increase in the number of autophagosomes upon AtEH1/Pan1 overexpression, the transcription of most ATG genes is upregulated in the cells transiently transfected with AtEH1/Pan1. The difference between two experiments was calculated by Student’s T-test, and the significance is indicated with a single (p ≤ 0.05) or double (P ≤ 0.01) asterisk. b, c Confocal images of root meristems (b) and root differentiation/maturation zone cells (c) of seedlings taken at the respective time points after transfer to β-Estradiol-containing medium. At 12 h post induction, clear cytoplasmic signal of AtEH1/Pan1 can be detected and the first autophagosomes appear in the differentiation/maturation zone. At 24 h post induction, the signal in the meristem cells is predominantly membrane-associated and autophagosomes in the differentiation/maturation zone are clearly present. At 48–72 h post induction, the signal in the meristem zone decreases, while autophagosomes remain very apparent in the differentiation/maturation zone (Scale bars: 15 μm). d After induction by transfer, AtEH1/Pan1 is detected after 12 h and increases up to 48 h. ATG8 protein levels also increase up to 48 h. ATG8 levels of transferred seedlings to non-induced and induced conditions are plotted and show a clear rise from 48 h onward, co-occurring with a decrease in AtEH1/Pan1 abundance. The plot shows the average of two biological repeats. Ponceau S staining was used as loading control. e Arabidopsis plants expressing pUBQ10::mCherry-ATG8e and AtEH1/Pan1-GFP, driven by the β-Estradiol inducible promoter. After induction, strong co-localization between the two proteins was found (Scale bars: 10 μm). f Absence of co-localization between induced AtEH1/Pan1-GFP punctae and the protein aggregation dye ProteoStat® shows that the AtEH1/Pan1-positive punctae are not aggresomes (Scale bars: 10 μm). g Silencing of AtEH1/Pan1 expression in Arabidopsis (two independent AtEH1/Pan1 RNAi lines, both of which with about 30% reduction in gene expression) significantly reduced the number of autophagosomes (visualized and measured by the amount of ATG8 positive punctae from an area of 20 × 50 μm, n = 10) upon nitrogen starvation, indicating that reduction of AtEH1/Pan1 affects autophagy activity (Scale bars: 10 μm). h. AtEH1/Pan1 RNAi plants exhibit retarded root growth (7.8 ± 1.1 mm and 8.0 ± 1.2 mm, respectively) in nutrient depleted medium (MS -N) compared to the wild type (10.1 ± 1.6 mm). To make each set of experiments comparable, the root length at stress conditions was normalized against its length measured at control conditions (n = 20). i. Quantitative Real-time PCR of ATG8 genes in AtEH1/Pan1 RNAi plants. The expression of most ATG genes is down-regulated in these plants which indicates a reduction in autophagic capacity. Letters indicate statistically different groups based on ANOVA analysis. Error bars are S.D., *P < 0.05, **P < 0.01, ***P < 0.001 in Student’s T-test