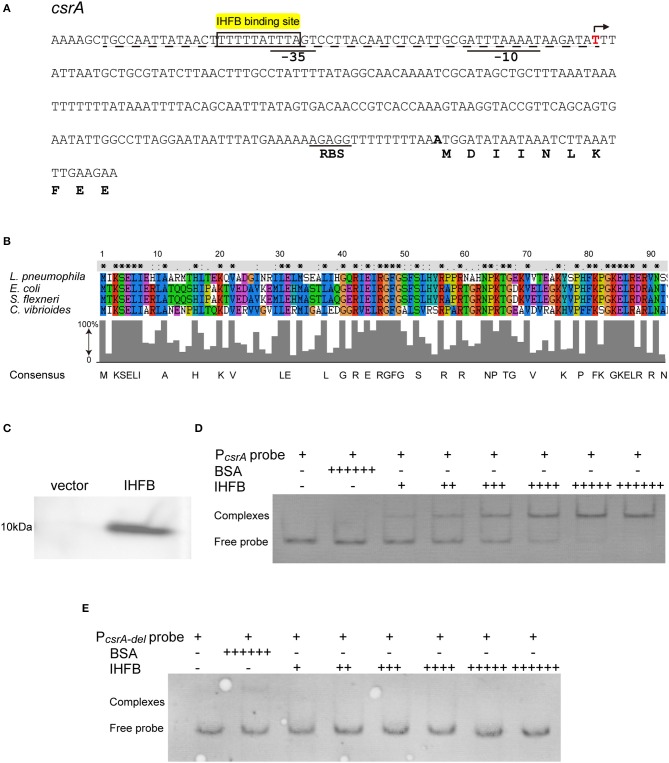
Figure 6.
IHFB binds directly to the regulatory region of csrA. (A) Map of the promoter regions of csrA and positions of putative IHF binding sites. The transcriptional start site is indicated by an angled arrow. Possible −10 site, −35 site, and RBS site sequences are underlined. Predicted IHFB binding sites are labeled in yellow. The dashed line indicates the 60 bp DNA fragment that is deleted in the “PcsrA-del” probe. Bioinformatics analysis was performed using Softberry software. (B) Sequence alignment of the putative IHFB from L. pneumophila with other prokaryotic IHF proteins. Numbers indicate the positions of amino acids in the sequences. Identical or similar residues are labeled with asterisks or periods, respectively. (C) Immunoblot of IHFB expressed in the BL21 strain of E. coli. The coding regions of IHFB were PCR amplified and fused to the expression vector pET-28a by the Gibson assembly method. The fusion gene constructs were transformed into E. coli strain BL21. Expression of IHFB was induced by IPTG to a final concentration of 10 μM. Proteins extracted from equivalent numbers of recombinant bacteria (1 × 109 cells) were loaded onto an SDS-PAGE gel and IHFB was detected using an anti-His tag antibody. (D,E) EMSA analysis of in vitro binding of IHFB to the promoter region of csrA (PcsrA probe) (D) and the promoter region deleted 60 bp upstream from the transcriptional start site of csrA (PcsrA-del probe) (E). An equivalent amount of the csrA probe DNA (200 ng) was added to every lane. BSA is used as a negative control for normalization, in which the amount is excessive (+ + + + ++). The first two lanes contain no IHFB (–) and the amount of IHFB is gradually increased (+). Each + sign indicates 200 ng of protein.