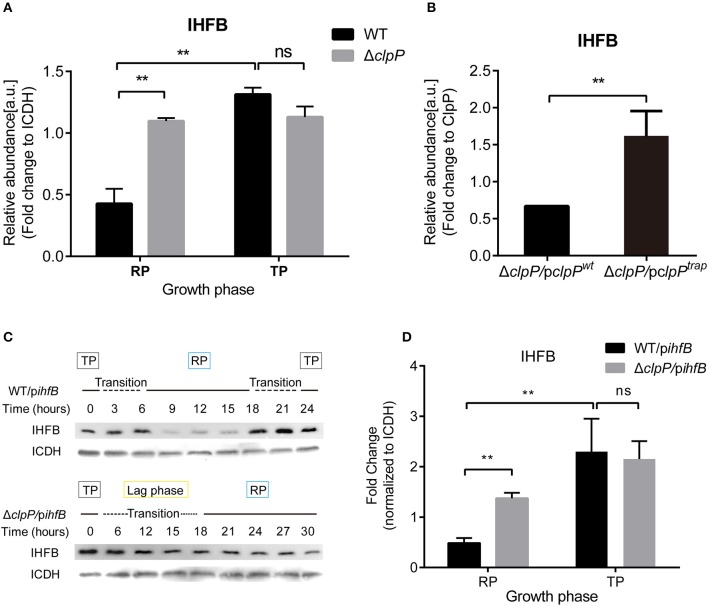
Figure 8.
IHFB is temporally expressed and is degraded by ClpP during the RP. (A) The abundance of IHFB in WT and ΔclpP determined by proteomics analysis. WT and ΔclpP were cultured in fresh AYE medium at the same initial OD600 values. Bacterial cells in the RP were harvested at an OD600 of 0.7–1.0 and those in the TP were harvested approximately 6 h after the cessation of growth. Total proteins from indicated samples were extracted for proteomic analysis. Bacterial whole-cell lysates from WT and ΔclpP were prepared and identified by mass spectrometry. ICDH was measured as a loading control. (B) The abundance of IHFB in ΔclpP/pclpPwt and ΔclpP/pclpPtrap were identified by ClpPtrap proteomic analysis. Bacterial cells in TP were harvested approximately 6 h after the cessation of growth. Whole-cell lysates from ΔclpP/pclpPwt and ΔclpP/pclpPtrap were prepared and His-tagged ClpP proteins were purified by Ni-NTA affinity. Substrates captured inside the proteolytic barrel were co-purified along with the His-tagged ClpP complex and identified by mass spectrometry. ClpP was measured as a loading control. (C) IHFB is degraded by ClpP during the RP. Whole-cell lysates were prepared from equal amounts of cells of WT/pihfB and ΔclpP/pihfB at indicated time points, and an immunoblot of IHFB was performed using an anti-His tag antibody. ICDH was probed as a loading control. (D) Relative protein levels of IHFB in the RP and the TP calculated by ImageJ. Bacterial cells in the RP were harvested at an OD600 of 0.7–1.0 and those in the TP were harvested approximately 6 h after the cessation of growth. The quantitative data were analyzed using two-way analysis of variance (ANOVA) test by GraphPad Prism. The values that are significantly different are indicated by a bar and asterisk as follows: **p < 0.01.