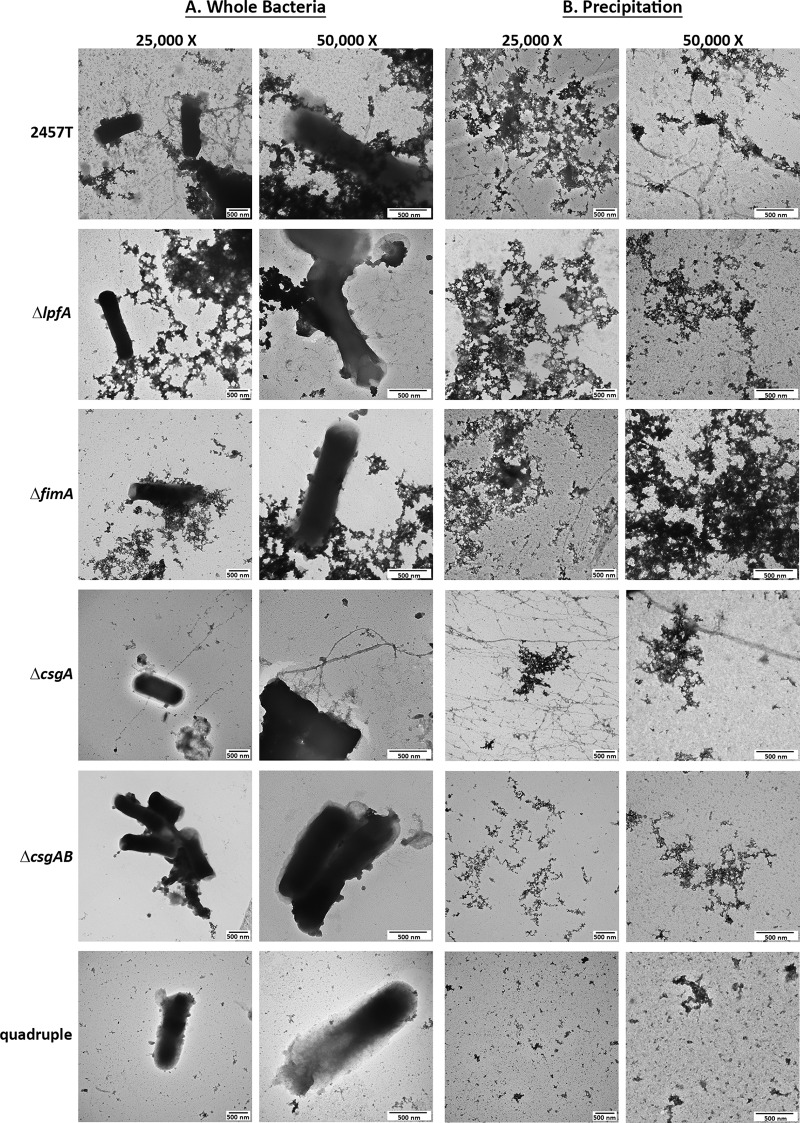
FIG 7.
TEM analysis for each S. flexneri 2457T adherence mutant. (A) The ΔlpfA, ΔfimA, ΔcsgA, ΔcsgAB, and quadruple mutants were grown statically overnight in IVLCs, subsequently processed for TEM analysis, and analyzed with wild-type strain 2457T as a control. Each mutation resulted in the loss of either the thicker structures (ΔlpfA mutant), the thinner structures (ΔfimA mutant), or the electron-dense aggregates (ΔcsgA and ΔcsgAB mutants). The quadruple mutant did not display visible structures. Images for wild-type strain 2457T are from experiments separate and biologically independent of those used to obtain the images provided in Fig. 1. (B) To verify the results, ammonium sulfate precipitation was performed to isolate and visualize structures from wild-type S. flexneri strain 2457T and each of the five mutants. The three types of factors can be visualized in wild-type bacteria; however, only two of the three structures were present for the single mutants. Each mutation resulted in the expected loss of structure, and no structures were visualized in the quadruple mutant. The data verify that the correct structural subunit was deleted for each mutant. All images are representative of those from at least two biological independent experiments. Different fields are presented for the images with ×25,000 and ×50,000 magnifications for all images in panels A and B, in which both sets of images display the 500-nm scale bar.