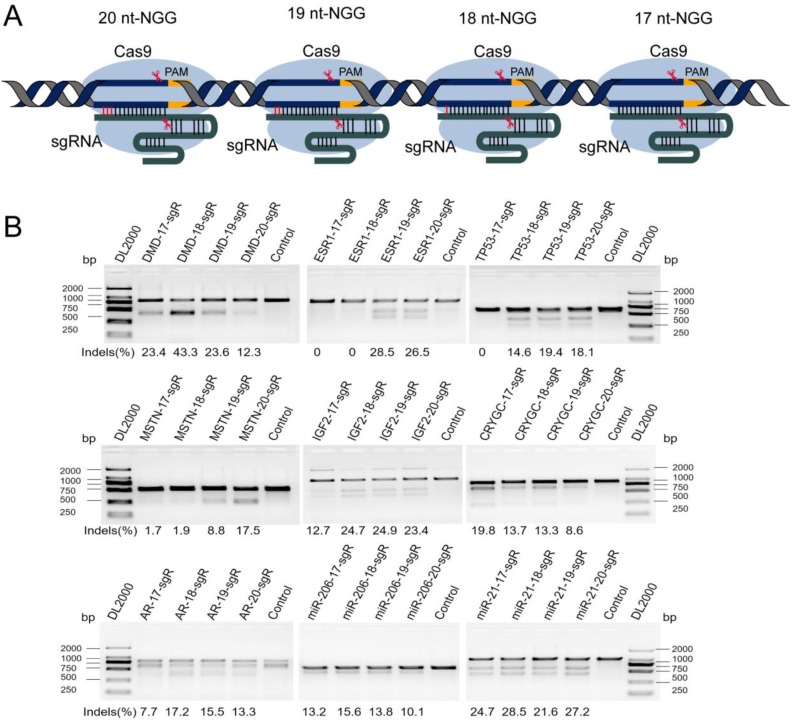
Figure 1.
Difference in the activity at the same genomic location in different lengths of sgRNAs. (A) Scheme of using 20, 19, 18, 17 nts of sgRNAs on target genes. The sequence patterns are recognized including N20NGG, N19NGG, N18NGG, and N17NGG. (B) Activities of sgRNAs in different lengths on target genes using T7ENI cleavage assay. “NGG” represents protospacer adjacent motif (PAM) sequences, N represents one of four bases, including adenine (A), guanine (G), cytosine (C), and thymine (T); sgR: single-guide RNAs; bp: base pairs; DL2000:DNA marker, control: wild-type control cells.