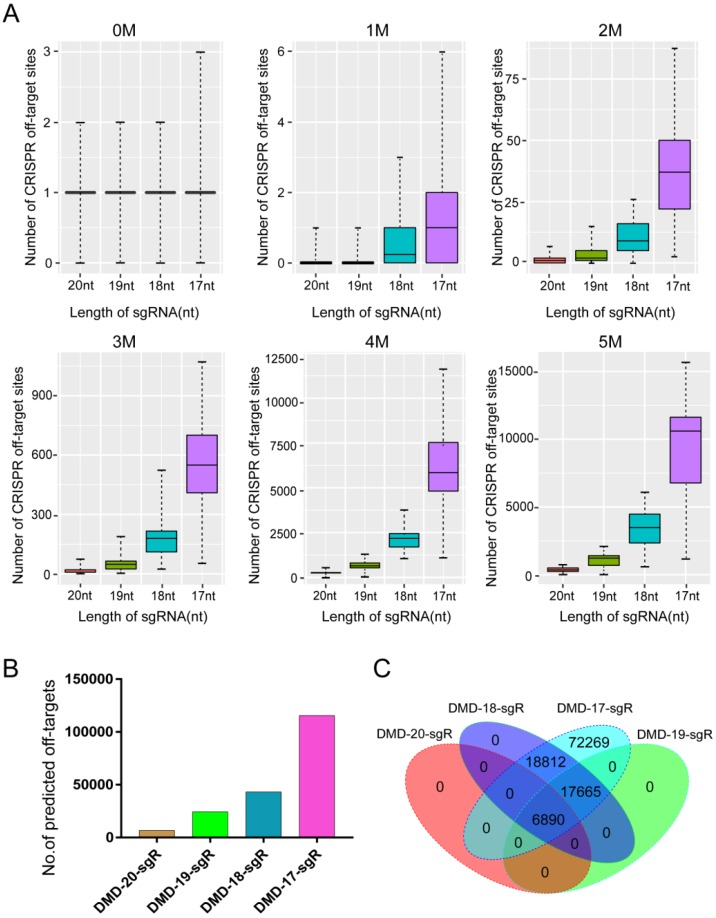
Figure 2.
Predicted specificity of sgRNAs in different lengths targeting DMD gene. (A) Differences in the predicted number of off-target sites with 1, 2, 3, 4, 5 nucleotide mismatches. T-test was performed on the total predicted off-target sites of sgRNAs with 19, 18, 17 nts against 20 nt in length. P values are 1.5e-11, 3.17e-16, 9.87e-17, respectively. (B) Difference in the predicted total number of off-target sites of 20, 19, 18, 17 nt sgRNAs. (C) Venn diagram of the predicted off-target sites in different lengths of sgRNAs. nt: nucleotides; sgR: small guide RNA; M representsthe number of nucleotide mismatches (1M, 2M, 3M, 4M, or 5M); 0M represents the perfect match to the on-target site; off-target sites are counted at the number of 0M sites > 1.