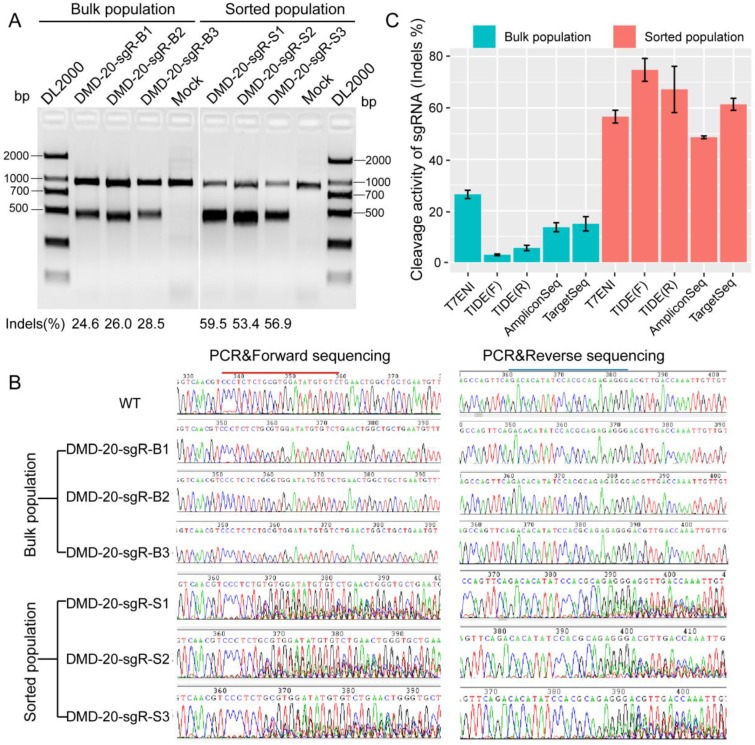
Figure 3.
The sensitivities of different methods in detecting the activity of 20 nt-long sgRNA. (A) The activity of sgRNA on targeting DMD gene by T7ENI cleavage assay. (B) PCR products were sequenced in both directions. (C) Assessing sgRNA activity by four methods. These methods includes T7ENI cleavage assay; Sanger DNA sequencing followed by TIDE web-based software analysis; High-throughput amplicon sequencing (AmpliconSeq); and Target capture sequencing (TargetSeq). Bulk population represents unsorted cells; Sorted population represents sorted cells; Mock control represents Lipofectamine 2000 only; WT: wild-type cells; TIDE(F): Sanger DNA forward-sequencing data; and TIDE(R): Sanger DNA reverse-sequencing data.