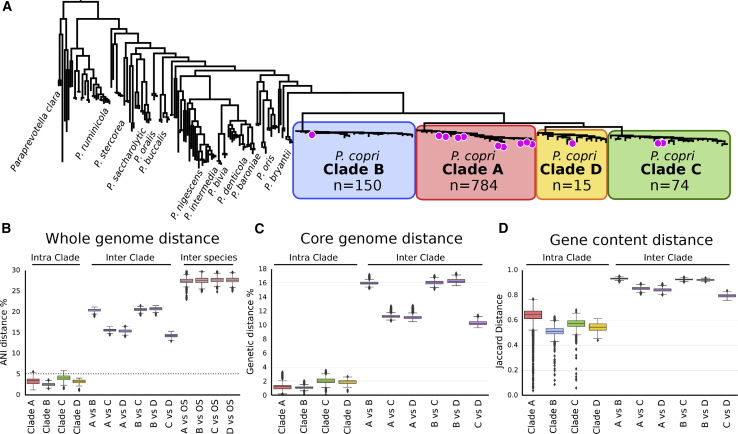
Figure 1.
The Four Distinct Clades of the P. copri Complex
(A) Whole-genome phylogenetic tree of a representative subset of the four P. copri clades comprising the P. copri complex in relation to other sequenced members of the genera Prevotella, Alloprevotella, and Paraprevotella. Magenta circles indicate P. copri isolate sequences (built using 400 universal bacterial marker gene sequences, see STAR Methods). The phylogeny containing all P. copri genomes is available as Figure S1B and http://segatalab.cibio.unitn.it/data/Pcopri_Tett_et_al.html (see Data and Code Availability; Method Details).
(B) Genetic distances within a clade (intra-clade), between clades (inter-clade), and between clades and other species (denoted as OS) of Prevotella, Alloprevotella, and Paraprevotella (inter-species), shown as pairwise average nucleotide identity distances (ANI distance). The dotted line denotes 5% ANI distance.
(C) Pairwise SNV distances based on core gene alignment within and between clades (see STAR methods).
(D) Jaccard distance based on pairwise gene content (see STAR Methods) between and within the P. copri clades.