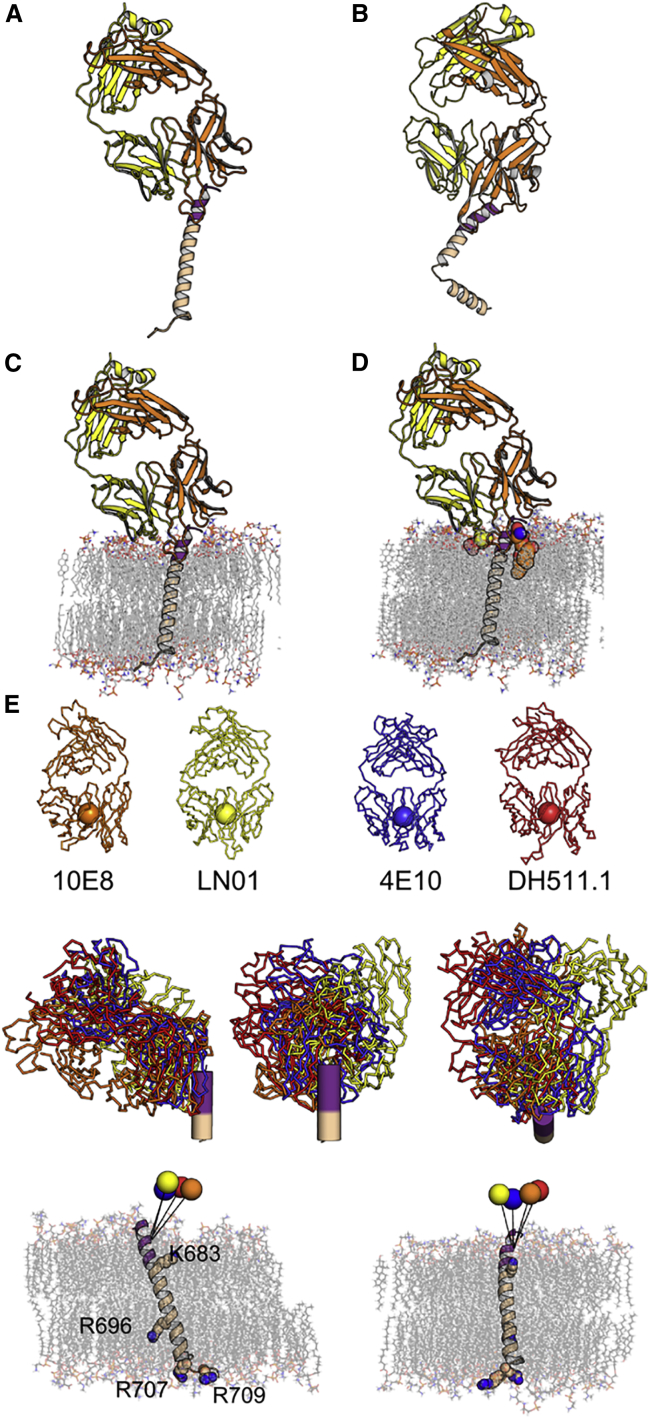
Figure 6.
Structure of LN01 in Complex with gp41 MPER-TM2 Reveals a Continuous Helix of MPER and TM
(A) Structure of the LN01-MPER-TM2 complex with the TM in the straight conformation. Color coding is the same as in Figure 5. Gp41 MPER residues 650 to 670 are disordered in the LN01-MPER-TM2 structure.
(B) Structure of the LN01-MPER-TM2 complex with the TM in the bent conformation.
(C) Orientation of the LN01-MPER-TM2 complex in the lipid bilayer based on the MD simulation result (lipids are represented in sticks).
(D) LN01-MPER-TM2 complex with modeled phosphatidylcholine and 06:0 PS (based on the LN01-MPER-TM1 structure) demonstrates that both lipids are well positioned to be part of the bilayer. Phosphatidylcholine and 06:0 PS are shown in spheres.
(E) The angle of approach of MPER bnAbs. The upper panel shows the bnAbs LN01 (yellow), 10E8 (orange), 4E10 (blue), and DH511.1 (red), represented with a sphere at the center of the variable domain for each antibody.
The middle panel shows their orientations upon recognition of the linear helical epitope as determined by Cα superposition of the MPER peptide of the four complexes (two side views and one top view looking down the helical axis of MPER).
The lower panel shows the representation of the angle of approach of the different bnAbs on the MPER-TM domain inserted in the lipid bilayer (lipids and basic residues are shown as sticks). The trajectory between the center of each antibody and gp41 T676 is depicted by a straight line.