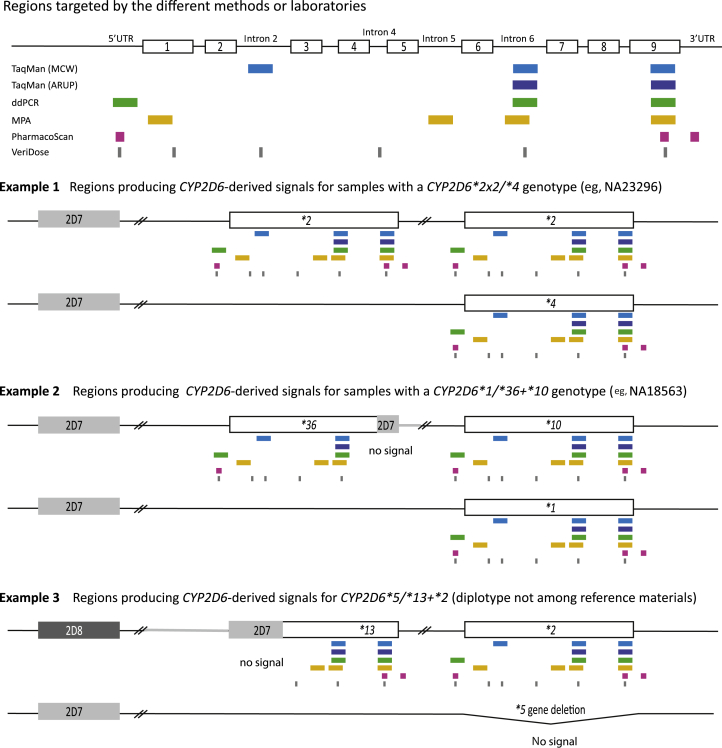
Figure 1.
Analysis of CYP2D6 copy number and structural variation. The figure provides an overview of the regions targeted for quantitative copy number determination by method and laboratory. The top panel shows the target regions of the TaqMan copy number assays used in the study. The Medical College of Wisconsin/RPRD Diagnostics (MCW) test site interrogated three gene regions whereas ARUP Laboratories (ARUP) tested for two gene regions. TaqMan assays were also used for droplet digital PCR (ddPCR) whereas a quantitative multiplex PCR method (MPA) developed at Children's Mercy Kansas City targets four regions. The PharmacoScan and VeriDose platforms target three and six regions, respectively. CYP2D6 exons are represented by numbered boxes and the 5′ untranslated region (5′UTR) and introns 2, 4, 5, and 6 also are shown. Example 1 shows a diplotype that produces the same copy number calls (three copies) across all targeted regions. Example 2 shows a diplotype containing an allele with a CYP2D7-derived exon 9 conversion; this region does not amplify and therefore produces two-copy number calls, whereas all other regions produce three-copy number calls. Example 3 shows a genotype with a CYP2D6*5 gene deletion on one chromosome and a tandem harboring a CYP2D7–CYP2D6 hybrid on the other chromosome. As illustrated, the 5′UTR, exon 1, and intron 2 target regions corresponding to CYP2D7 do not amplify and produce one-copy number calls, whereas the intron 5 and 6 and exon 9 regions amplify from the CYP2D6 portion of the hybrid as well as the CYP2D6*2 in the tandem producing two-copy number calls. Table 1 summarizes the copy number calls for each example for each of the methods/platforms used.