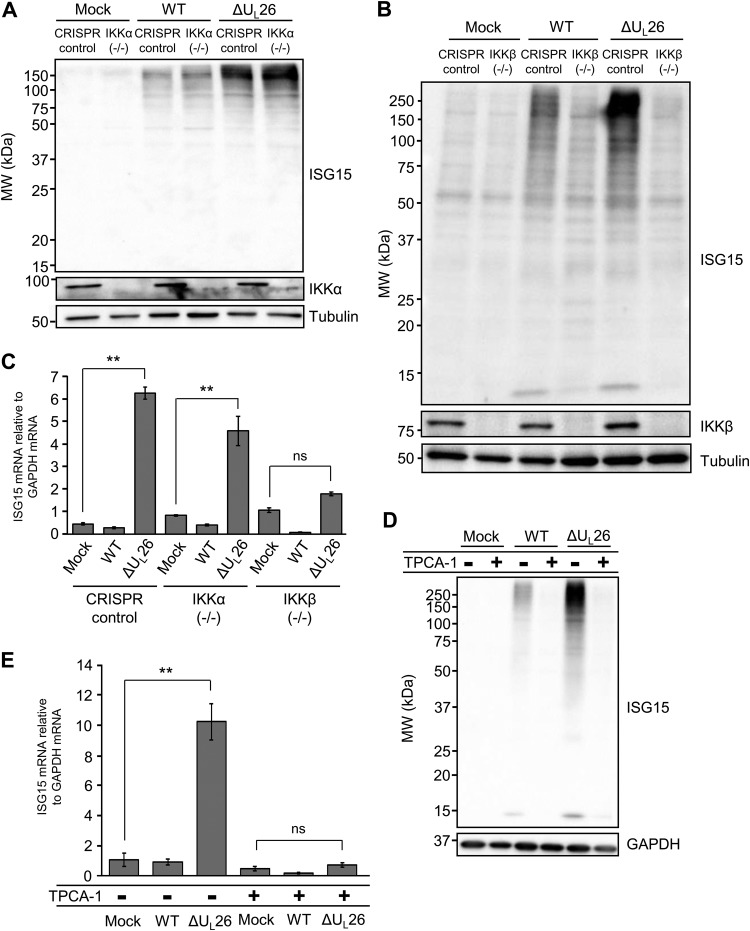
FIG 6.
Hyper-ISGylation during ΔUL26 mutant infection requires IKKβ. (A) CRISPR control and IKKα(−/−) BJ/hTert cells were either mock infected or infected with WT or ΔUL26 mutant HCMV at an MOI of 3.0. Cell lysates were harvested at 48 hpi and Western blotted with the indicated antibodies. (B) CRISPR control and IKKβ(−/−) BJ/hTert cells were infected as in panel A, and cell lysates were harvested at 48 hpi for Western blotting with the indicated antibodies. (C) Total cellular RNA was harvested at 48 hpi from CRISPR control, monoclonal IKKα(−/−), and a pooled polyclonal population of IKKβ(−/−) BJ/hTert cells infected as in panel A. The abundance of ISG15 mRNA was measured by real-time PCR and normalized to GAPDH. Values are the means ± SEM (n = 8; **, P < 0.01; ns, not significant). (D) MRC5 fibroblasts were pretreated with 10 μM TPCA-1 for 4 h prior to either mock infection or infection with WT or ΔUL26 mutant HCMV at an MOI of 3.0. Cell lysates were collected at 48 hpi for Western blotting with the indicated antibodies. (E) MRC5 cells were pretreated with TPCA-1 and infected as in panel D. Cellular RNA was harvested at 48 hpi, and the accumulation of ISG15 mRNA was measured by real-time PCR and normalized to GAPDH. Values are plotted as the means ± SEM (n = 5; **, P < 0.01; ns, not significant).