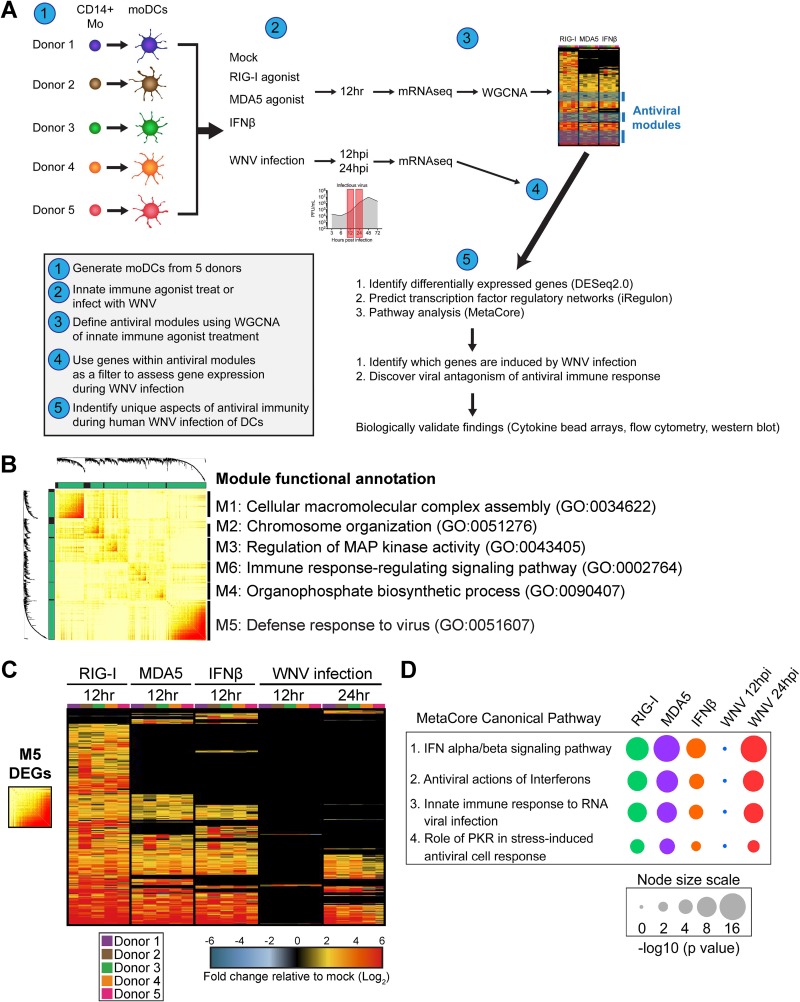
FIG 3.
Systems biology approach reveals antiviral landscape downstream of RLR signaling and WNV infection. (A) Overview of systems biology approach used in this study. (B) Topologic overlap matrix showing enriched modules defined by WGCNA following a 12-h treatment with RIG-I agonist (100 ng/1e6 cells), MDA5 agonist (100 ng/1e6 cells), or IFN-β (1,000 IU/ml). Functional annotation was performed using DAVID Bioinformatics Resource, version 6.8, with the top enriched biological process shown. GO, Gene Ontology category. (C) Heat map of all module 5 differentially expressed genes with the log2 normalized fold change relative to levels in uninfected, untreated cells shown. Genes that did not reach the significance threshold are depicted in black. (D) Top enriched MetaCore canonical pathways of module 5 differentially expressed genes relative to expression in uninfected and untreated cells (>2-fold change; P < 0.01). Node size corresponds with the pathway enrichment significance score (–log10 P value) for each indicated treatment condition.