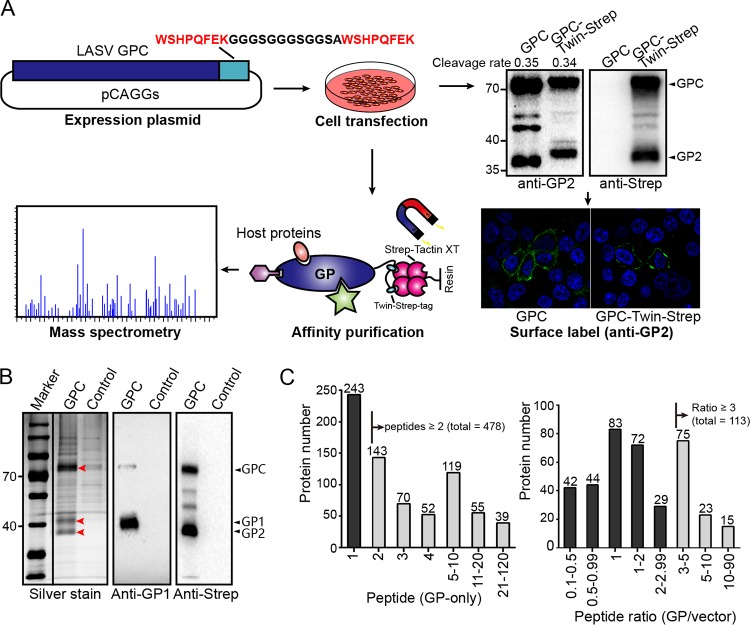
FIG 1.
Identification of host proteins interacting with LASV GP. (A) Schematic diagram of the experimental strategy used to identify host proteins interacting with LASV GP. HEK293T cells were transfected with a pCAGGs plasmid encoding LASV GP with a C-terminal Twin-Strep-tag or a pCAGGs plasmid expressing only the Twin-Strep-tag as a control. At 48 h posttransfection, the cells were harvested and lysed by using a mild nonionic detergent buffer at 4°C. LASV GP-associated host interactors were pulled down from cell lysates by magnetic Sepharose beads coated with Strep-Tactin XT. Purified proteins were digested into peptides by trypsin and subjected to liquid chromatography-tandem mass spectrometry analysis. Western blotting and immunofluorescence were used to test whether the Twin-Strep-tag would affect the proteolytic cleavage, glycosylation, and cell surface transport of LASV GP. The cleavage rates of tagged and untagged GP2 are shown above, as determined by densitometric quantification. (B) Silver-stained SDS-PAGE gel of purified LASV GP together with host interactors or the control group. Western blot analysis was also conducted to show the bands of viral GP1 and GP2, as detected by GP1-specific serum and an anti-Strep antibody, respectively. The silver-stained and Western blot gels shown here are representatives of three independent replicates. (C) The left column diagram shows the distribution of detected peptide counts from proteins identified in only the GP group and not in the control group. The right column diagram shows the distribution of peptide count ratios from proteins identified in both the GP group and the control group (GP/control). Proteins that fulfilled our filtration standard in the two groups were considered to be valid interactors.