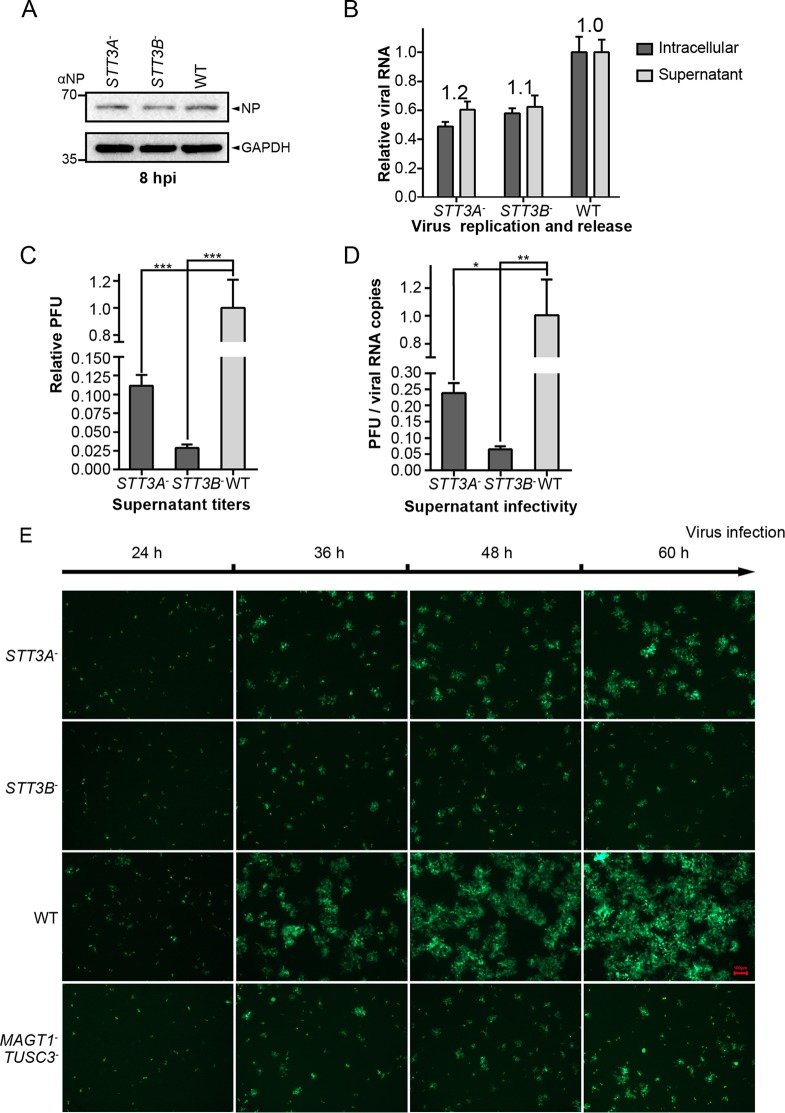
FIG 5.
Knockout of STT3A or STT3B led to the formation of viral particles with reduced infectivity. (A) Single-step infection assay was performed to assess the productive entry and early infection of the rLCMV/LASV GPC virus in STT3A– and STT3B– cells. The cells were infected with virus at an MOI of 0.01, incubated for 30 min at 4°C to synchronize the entry process, and then shifted to 37°C to allow penetration. Cells were harvested at 8 hpi, and the viral NP level was detected by Western blotting with anti-NP serum. (B) Intracellular and supernatant genome virus levels were measured at 48 hpi to assess virus budding ability by qRT-PCR. The ratio of the supernatant viral genome level versus the intracellular viral genome level was calculated as an indicator of virus budding ability (presented above the column). (C) The relative PFU in the supernatants of different cells at 48 hpi are shown. (D) The ratios of viral PFU versus viral genome RNA copy numbers were calculated as an indicator of viral infectivity. All data displayed here represent the means ± the standard deviations (SD) of three independent experiments, and each independent experiment had two replicates. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (E) Viral foci were observed by fluorescence microscopy at different time points in cells infected with the rLCMV (NP-P2A-GFP)/LASV GPC virus. The foci in STT3A– and STT3B– cells were notably smaller than those in WT cells. All of the micrographs displayed here were collected at the same scale (the length of the red bar represents 100 μm) and are representatives of several images.