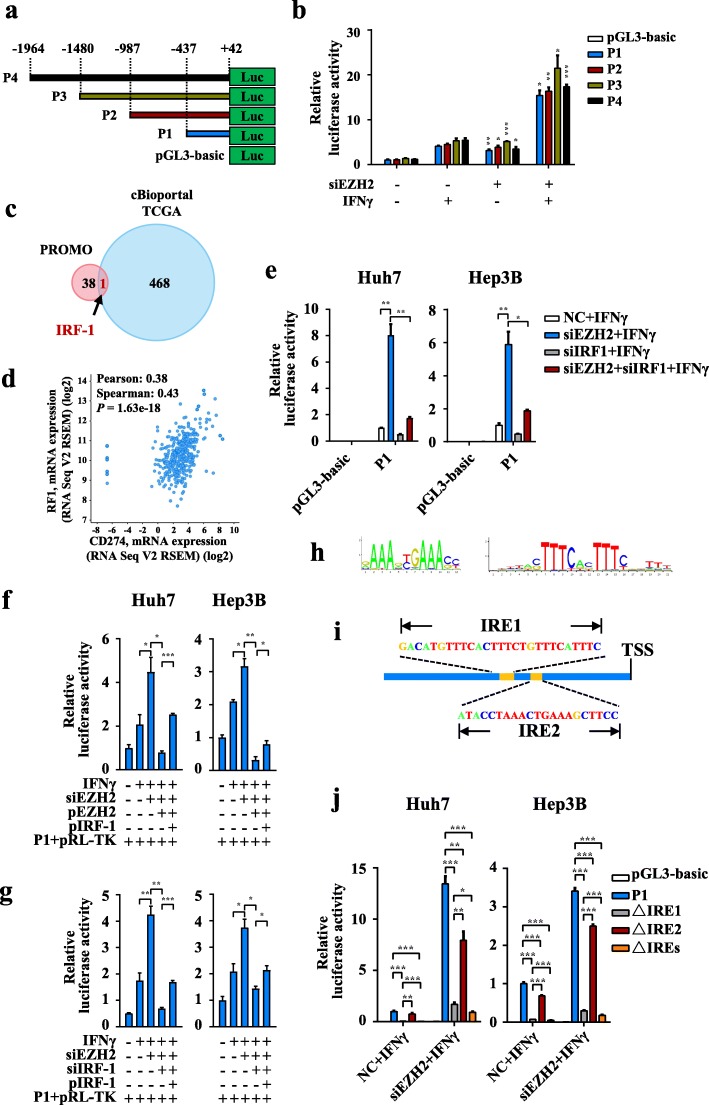
Fig. 3.
IRF1 is a potential transcription factor involved in the negative regulation of PD-L1 by EZH2. a Schematic diagram of a series of CD274 (PD-L1) gene promoter luciferase reporter plasmids. b After transfection with EZH2-targeted or NC siRNA overnight, Huh7 cells were co-transfected with pGL3-basic vector or the indicated CD274 promoter luciferase reporter gene plasmid and the pRL-TK plasmid for 48 h, and then treated with IFNγ for an additional 24-h. Luciferase activity was determined and normalized using the dual luciferase reporter system. (Mean ± S.E.M.; n = 3; the asterisk represents a comparison between the siEZH2 group and the corresponding control group; * P < 0.05, ** P < 0.01, *** P < 0.001, Wilcoxon test). c TFs that could potentially bind to the P1 truncated promoter were predicted using the PROMO bioinformatics software (pink circle). Genes showing the absolute values of both the Pearson and Spearman expression correlation coefficient (positively or negatively correlated) of more than 0.3 with CD274 (PD-L1 gene) in HCC tissues (TCGA, Provisional) were analyzed on the cBioportal website (blue circle). Venn diagram showing that IRF-1 was the only candidate gene in both gene sets. d Scatter gram showing the mRNA expression correlation of CD274 and IRF1 from TCGA (HCC, Provisional). Pearson and Spearman correlation coefficients and P values are shown. e Huh7 and Hep3B cells were transfected with NC or EZH2-targeted, IRF1-targeted, or both, siRNA overnight, and then co-transfected with pGL3-basic vector or the P1 luciferase reporter gene plasmid and pRL-TK plasmid for 48 h. The cells were then treated with IFNγ for an additional 24 h. Luciferase activity was determined and normalized using the dual luciferase reporter system (Mean ± S.E.M.; n = 4; * P < 0.05, ** P < 0.01, Wilcoxon test). f After transfection with NC or EZH2 siRNA targeting 3′-UTR, Huh7 and Hep3B cells were transfected with the indicated plasmids for 48 h, and then treated with IFNγ for 24 h. Luciferase activity was determined and normalized using the dual luciferase reporter system (Mean ± S.E.M.; n = 3; * P < 0.05, ** P < 0.01, Wilcoxon test). pEZH2 and pIRF-1 represent ectopic expression of EZH2 and IRF-1 respectively, and the corresponding control groups were transfected with NC siRNA and/or vector plasmids. g After transfection with NC or the indicated siRNA targeting 3′-UTR, Huh7 and Hep3B cells were transfected with the indicated plasmids for 48 h, and then treated with IFNγ for 24 h. Luciferase activity was determined and normalized using the dual luciferase reporter system (Mean ± S.E.M.; n = 3; * P < 0.05, ** P < 0.01, Wilcoxon test). pIRF-1 represent ectopic expression of IRF-1, and the corresponding control groups were transfected with NC siRNA and/or vector plasmids. h Sequence logo of IRF1 binding site frequency matrix of Homo sapiens predicted using the online software JASPAR. i Schematic representation of IRF1 binding sites in the CD274 P1 promoter region, as predicted by JASPAR. IRE, IRF1 response element. j Huh7 and Hep3B cells were transfected with NC or EZH2-targeted siRNA overnight, and then co-transfected with pGL3-basic vector or the indicated P1 with or without IREs sequence deletion luciferase reporter gene plasmid and pRL-TK plasmid for 48 h. The cells were then treated with IFNγ for an additional 24 h. Luciferase activity was determined and normalized using the dual luciferase reporter system (Mean ± S.E.M.; n = 4; NS, no significant difference; * P < 0.05, ** P < 0.01, *** P < 0.001, Wilcoxon test)