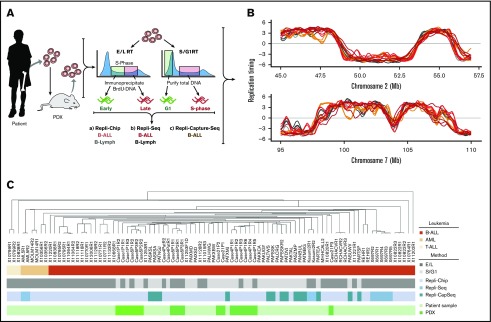
Figure 2.
Methods for genome-wide RT profiling. (A) Bone marrow samples were collected from pediatric BCP-ALL patients and analyzed by 1 of the methods shown. Samples with high viability were analyzed by early/late (E/L) RT methods, and samples with poor viability were analyzed either by S/G1 methods or they were expanded in patient-derived xenografts (PDXs) to rejuvenate their viability for analysis by E/L RT methods.14 RT was measured using either E/L or S/G1 methods.14,28 In the E/L method, asynchronously growing cells are pulse-labeled with 5′-bromo-2′-deoxyuridine (BrdU) and separated by FACS into early and late S phase populations. BrdU-substituted DNA is immunoprecipitated with an anti-BrdU antibody and subjected to either microarray analysis (E/L Repli-ChIP16 or NextGen sequencing [E/L Repli-seq17]) to obtain an RT ratio [= log2(early/late)] along each chromosome. For the S/G1 method, unlabeled cells are sorted into S and G1 phase fractions, the relative abundance of genomic sequences is quantified by either microarray (S/G1 Repli-ChIP) or evenly spaced segments of genomic DNA are captured using oligonucleotides and subject to NGS (Repli-capture-seq). (B) Exemplary RT profiles showing the similarity in results using the various methods after appropriate local polynomial smoothing (Loess). Repli-Chip normal B-cell sanples are shown in grey, Repli-Chip B-ALL in orange, Repli-seq normal B-cells in black, Repli-seq B-ALL in red and Repli-Capture-seq in dark red. (C) Hierarchical clustering analysis of all patient data sets based on their genome-wide RT programs confirms that there is no bias from the different methods exploited for RT profiling.