Abstract
Background
Increasing evidence has suggested that gut flora play an important role in tumor progression and prognosis. However, the relationship between fecal microbiota and hematologic malignancy requires further investigation. This study aimed to characterize the relationship of the fecal microbial community in multiple myeloma (MM) patients.
Material/Methods
A total of 40 MM patients and healthy controls (n=17) were retrospectively collected from the First Affiliated Hospital of Sun Yat-sen University between October 2018 and May 2019. The fecal samples were collected for 16S rRNA high-throughput sequencing for the fecal microbial community, as well as diversity and correlation analysis. Furthermore, 21 MM patients and their family members were selected for the matched pair analysis to confirm the fecal microbiota taxonomic changes by qRT-PCR assay.
Results
Diversity analysis showed that diversity measured by Shannon index was lower in MM patients compared with healthy controls. At the phylum level, higher abundances of Proteobacteria but lower abundances of Actinobacteria were identified in the MM group in comparison with the healthy control group. At the genus level, the proportion of Bacteroides, Faecalibacterium, and Roseburia was significantly higher in the MM group. The matched pair analysis showed that Pseudomonas aeruginosa and Faecalibacterium were significantly more abundant in the MM group. Further analysis on prognostic risk factors revealed that the Faecalibacterium prausnitzii level was significantly correlated with ISS stage.
Conclusions
Our study highlights the imbalanced composition and diversity of the gastrointestinal microbiome in MM patients, which could be further used as a potential biomarker for MM risk screening, therapeutic strategies, and prognostic prediction.
MeSH Keywords: High-Throughput Nucleotide Sequencing, Microbiota, Multiple Myeloma, Prognosis
Background
Multiple myeloma (MM) is a neoplastic disorder that is characterized by the infiltration and clonal proliferation of antibody-secreting post-germinal center plasma cells (PCs) in the bone marrow (BM) [1]. Although several risk factors are associated with MM, such as life style, genes, dietary, occupation, environment and, especially, infection [2,3], the etiology and pathogenesis remain largely undefined.
The intestinal microbiota (IM) is an ecosystem consisting of more than 1013 microorganisms, including the symbiotic microorganisms resident in the digestive tract and exogenous microorganisms quickly passing through the digestive tract [4]. The microbiota in the human intestinal tract not only participates in the metabolic absorption of nutrients in the host intestinal tract (such as short-chain fatty acids and nitrogen-containing compounds) [5], but also regulate the internal intestinal environment by secreting substances such as butyric acid [6]. Substances secreted by the gut microbiota, such as lipopolysaccharide (LPS), or components of the bacteria themselves, such as flagellin, can bind to pattern recognition receptors (PRRs) in human intestinal mucosal epithelial cells to initiate downstream signal transduction pathways, thereby regulating cytokine secretion in epithelial cells [7,8]. After entry into the circulatory system, these cytokines can affect hepatic and cerebral functions through the nervous immune system, leading to corresponding organ metabolism disorders and diseases [9].
Studies have confirmed the relationship between IM disorders and tumorigenesis and tumor progression. For example, Escherichia coli, Clostridium, and enterotoxin-producing Bacteroides fragilis can trigger colon cancer [10,11]. Alteration of microbiota can alter many signals between the colonizing bacteria and epithelial or immune cells, leading to changes in inflammation, epithelial cell cycle, proliferation, or mucus production. Some of these changes promote cell transformation or DNA damage, which are risk factors for developing precancerous lesions and cancer [12]. It has also been reported that the Helicobacter pylori level in the biliary tract is closely correlated with extrahepatic cholangiocarcinoma [13]. The imbalanced IM induced by the lower immune function can directly cause bacterial infection, and the products, toxins, and metabolites of the IM (such as short-chain fatty acids) can enter the mesenteric lymph nodes through the intestinal wall to further enter the circulatory system, thereby stimulating systemic immune response [14]. Under the synthetic interaction of innate and adaptive immune cell migration, cytokines, endocrine, and nervous system, IM can also affect other organs of the host that are involved in the pathogenesis of various cancers, including breast cancer, liver cancer, pancreatic cancer, and other tumors [15–17].
A previous study has demonstrated the relationship between the pathogenesis of hematologic malignancy and microbiota, mainly in acute lymphoblastic leukemia (ALL) [18]. However, the relationship between fecal microbiota and MM incidence is still unknown. The current study was designed to characterize the fecal microbial community in MM patients and to evaluate the relationship between fecal microbes and MM.
Material and Methods
Patients
A total of 40 MM patients were enrolled from Oct 2018 to May 2019, who were diagnosed according to IMWG criteria [19] and WHO classification [20]. Healthy controls were recruited from among the healthy spouses, children, and parents of the MM patients, who lived together with patients, had the same eating and living habits, had healthy reports in past medical history, and had no history of acute or chronic diseases. The exclusion criteria were: (1) Patients with other diseases that have been validated to affect the IM, including digestive diseases such as liver cirrhosis, liver cancer, inflammatory bowel disease, and irritable bowel syndrome; systemic diseases such as diabetes and hypertension; and thyroid diseases. (2) Patients treated with antibiotics, chemotherapy, plasma exchange, or bone marrow transplantation; subjects having cold, fever or other infection within 3 months before sampling, those administered antibacterial drugs, gastrointestinal motility drugs, or micro-ecological conditioning agents, or those having dramatic changes in eating and living habits 1 week before sampling. (3) Women in the menstrual period, or under with special conditions such as abdominal pain, diarrhea, and pregnancy.
This study was approved by the Ethics Committee of the First Affiliated Hospital of Sun Yat-sen University (2018201) and written informed consents were signed by all patients in accordance with the Declaration of Helsinki. This study was registered with the Chinese Clinical Test Registration Center, registration number ChiCTR1800019153.
Sample collection, storage, and preparation
A stool sample of each patient enrolled in this study was collected before any anti-myeloma chemotherapy and anti-infection therapies. Before fecal sample collection, patients were asked to urinate as much as possible to avoid urinary contamination. A disposable spoon was used to select the middle section of the feces, which was further placed in the sampling tube. The sampling time was no more than 30 min.
Samples were frozen in liquid nitrogen within 2 h after sampling. In brief, the frozen stool sample was cut into 400 mg/each by using a sterile scalpel, which was then put into a 2-mL sterile centrifuge tube. We extracted 3–6 samples and stored them at −70°C.
DNA extraction
Mobio PowerFecal® DNA Isolation kits were purchased to extract DNA from the stool sample in each group. Briefly, after adding 400 mg stool sample into the bead tube, 750 μL BeadSolution was further added to homogenize the sample for suspension, followed by addition of 60 μl of C1 solution. After mixing and inverting, the samples were placed in a 65°C water bath for 10 min, vortexed for 10 min, and centrifuged at 14 000 g for 2 min. Afterwards, 550 μL of supernatant was transferred to a new centrifuge tube, followed by addition of 250 μL of C2 solution, incubation in a 4°C ice bath for 5 min, and centrifugation at 14 000 g for 2 min. The procedure was performed in accordance with the manufacturers’ instructions. The finally obtained DNA was dissolved in 120 μl of C6 solution. DNA concentration of the obtained flora DNA was measured by an ultraviolet spectrophotometer NanoND-1000. In addition, primers 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and 806R (5′-GGTGGGTTATGGTCTTCAAAAGG-3′) of the 2 hypervariable regions V3+V4 of the 16sRNA gene were used for amplification, followed by electrophoresis in 1.8% agarose and determination of the extraction effect.
16s rRNA high-throughput sequencing
Thermocycling consisted of a 5-min denaturation at 94°C, 30 cycles of 98°C (30 s), annealing for 30 s (57–79°C), and 72°C for 2 min, and a final extension at 72°C for 10 min, followed by collection of a 5-μL sample for sequencing. Paired-end sequencing using the Illumina Hiseq sequencing platform was performed. The original data file was transformed into the original sequencing sequence by clip recognition analysis. The minimum overlap length between each sequence was 10 bp and the maximum mismatch ratio was set at 0.2; therefore, paired-end sequence data obtained from Hiseq sequencing were merged into 1 tag, simultaneously followed by quality-controlled filtering of the quality of the readable area (Reads) and the merging effect. FLASH v1.2.7 software was used to merge the Reads of each sample, which were Raw Tags. Trimmomatic v0.33 software was used to filter the merged Raw Tags to obtain high-quality Tags data (Clean Tags). Then, UCHIME v4.2 software was used to identify and remove chimeric sequences to obtain the final valid data (Effective Tags). According to the similarity level of 97%, the sequence was clustered by QIIME software, operational taxonomic unit (OTU) was categorized, followed by differential analysis on sample composition classification, abundance, and inter-sample difference analysis.
Quantitative real-time PCR (qRT-PCR)
We further validated the IM from 21 pairs of patients and their relatives using qRT-PCR. The conventional PCR product of the target microbiota gene was used as a standard control, followed by amplification using the TaKaRa® PremixTaq kit according to the manufacturers’ instructions. Three-step PCR reaction was used for amplification, and the primer sequences are shown in Table 1. The reaction conditions were as follows: pre-denaturation at 94°C for 5 min, denaturation at 98°C for 30 s, annealing for 30 s, extension at 72°C for 2 min (30 cycles), and final extension at 72°C for 10 min.
Table 1.
Primer sequence for the target flora detecting.
| Target flora | Primer name | Sequence (5′-3′) | Annealing temperature (°C) |
|---|---|---|---|
| Enterococcus faecails | Enco-F | AACCTACCCATCAGAGGG | 57 |
| Enco-R | GACGTTCAGTTACTAACG | ||
|
| |||
| Lactobacillusgroup | Lac-F | AGCAGTAGGGAATCTTCCA | 58 |
| Lac-R | ATTYCACCGCTACACATG | ||
|
| |||
| Clostridium cluster XI | CXI-F | ACGCTACTTGAGGAGGA | 58 |
| CXI-R | GAGCCGTAGCCTTTCACT | ||
|
| |||
| Faecalibacteriumprausnitzii | Pfra-F | GATGGCCTCGCGTCCGATTAG | 58 |
| Pfra-R | CCGAAGACCTTCTTCCTCC | ||
|
| |||
| Pseudomonas alcaligenes | Psd-F | CAAAACTACTGAGCTAGAGTACG | 60 |
| Psd-R | TAAGATCTCAAGGATCCCAACGGCT | ||
|
| |||
| Pseudomonasaeruginosa | Ps2-F | CCTGACCATCCGTCGCCACAAC | 79 |
| Ps2-R | CGCAGCAGGATGCCACGCC | ||
|
| |||
| Gram-positivebacteria | Gp-F | GAYGACGTCAARTCMTCATGC | 69 |
| Gp-R | AGGAGGTGATCCAACCGCA | ||
|
| |||
| Gram-negativebacteria | Gn-F | AYGACGTCAAGTCMTCATGG | 52 |
| Gn-R | AGGAGGTGATCCAACCGCA | ||
Afterwards, 5 μL of the reaction product was subjected to 1.5% agarose gel electrophoresis to examine the PCR product. The agarose gel of the target gene product was purified using an Ultra-Clean® 15 Nucleic Acid Purification Kit (Mobio), followed by determination of the purified DNA concentration by an ultraviolet spectrophotometer NanoND-1000 according to the formula:
The copy number of the target DNA per μL of solution was calculated (Table 2).
Table 2.
Target gene length and copy number of each standard sample (1 μL).
| Target flora | Target segment length (bp) | Initial copy number (1×1011) |
|---|---|---|
| Enterococcus faecalis | 357 | 1.54 |
| Lactobacillus group | 341 | 3.29 |
| Clostridium cluster XI | 135 | 6.90 |
| Faecalibacterium prausnitzii | 198 | 5.09 |
| Pseudomonas alcaligenes | 205 | 3.52 |
| Pseudomonas aeruginosa | 295 | 2.65 |
| Gram-positive bacteria | 354 | 2.80 |
| Gram-negative bacteria | 334 | 2.62 |
After the purified DNA was diluted to 1×108 copies/μL, 8 concentration gradients were further set by dilution at 1: 10 for a final concentration of 1×108, 1×107, 1×106, 1×105, 1×104, 1000, and 100 copies/μL of the standard solution. SYBR® Green qRT-PCR was further used to determine the Ct value, along with plotting a standard curve to mimic the standard equations of each target microbiota (Table 3).
Table 3.
Standard curve equation of each target flora.
| Target flora | Standard curve equation | R2 |
|---|---|---|
| Enterococcus faecalis | Y=−3.871X+37.950 | 0.992 |
| Lactobacillus group | Y=−3.939X+38.501 | 0.986 |
| Clostridium cluster XI | Y=−3.130X+35.783 | 0.978 |
| Faecalibacterium prausnitzii | Y=−3.460X+33.788 | 0.997 |
| Pseudomonas alcaligenes | Y=−3.098X+32.045 | 0.955 |
| Pseudomonas aeruginosa | Y=−3.743X+36.322 | 0.996 |
| Gram-positive bacteria | Y=−3.687X+35.916 | 0.995 |
| Gram-negative bacteria | Y=−3.240X+31.931 | 0.997 |
The initial copy number of the target gene in the sample was calculated against the standard curve, and the number of intestinal bacteria in the sample was absolutely quantified to calculate the initial copy number of genes per stool sample (40 mg).
Statistical analysis
The baseline clinical data of patients at admission are shown as mean±standard deviation. SPSS ver 20.0 was used for statistics. The unpaired t test was used for analysis, and the chi-square test was used to compare sex distributions. Alpha diversity analysis was performed using Chao1 index and Shannon index, and Beta diversity analysis was performed by PCoA principal coordinate analysis in the 16s rRNA high-throughput sequencing data analysis. LEfSE and Metastats were used for significance analysis between groups. PICRUSt software was used for functional gene prediction. P<0.05 was considered as statistically significant.
Result
Comparison of clinical baseline data of patients between groups
The baseline data of patients at admission for each group of were shown in Table 4. There were no statistical differences in age, gender, BMI index, creatinine, and albumin levels. In addition, patients’ serum glucose level or HbA1c levels were also detected. Patients were divided into 2 groups (ISS I–II and ISS III), and there was no significant difference in serum glucose level (5.05±0.99 g/L and 5.08±0.75 g/L, P=0.91) and HbA1c levels (5.79±0.72% and 5.88±0.46%, p=0.66) between the 2 groups.
Table 4.
Baseline data of subjects.
| MM (n=40) | Healthy control (n=17) | P value | |
|---|---|---|---|
| Age | 56.53±9.31 | 50.18±12.17 | 0.141 |
| Gender (M/F) | 21/19 | 7/10 | 0.434 |
| BMI | 23.02±3.22 | 23.08±1.87 | 0.942 |
| Albumin (g/L) | 33.17±7.00 | – | |
| Creatinine (umol/L) | 105.93±65.87 | – | |
| ISS | I–II (21) III (19) |
||
| Subtype | IgG-κ (10) IgG-λ (7) IgA-κ (7) IgA-λ (5) IgD-λ (3) κlight chain (3) λlight chain (5) |
Decreased fecal microbiota diversity in patients with MM
The species diversity within a single sample was reflected by Alpha diversity. In this study, Chao1 index and the Shannon index were used to measure the Alpha diversity of the sample. Chao1 index is used to measure the number of species within the community, and Shannon index is used to measure the abundance of the community, which is affected by species abundance and uniformity in the community. Larger Chao1 and Shannon indexes indicate the higher diversity of the sample species. The sample diversity index is shown in Table 5. A higher coverage value of the sample OUT suggests a better representativeness. The OTU coverage of both groups exceeded 99.9%, suggesting that the classification results of the sequencing data were reliable. Compared with the healthy control group, the OTU number (P=0.295) and the Chao1 index were both decreased (P=0.332) in the MM group, but without statistical significance. Shannon index was decreased (P<0.05), indicating the number of species was not significantly decreased, but the uniformity of the species was decreased in the IM of MM patients, suggesting structural change of IM in MM patients, and decreased diversity of uniformity in IM.
Table 5.
Alpha diversity index in each group.
| Healthy controls (n=17) | MM (n=40) | P value | |
|---|---|---|---|
| OUT value | 201.76±62.07 | 184.1±55.82 | 0.295 |
| Chao1 Index | 233.79±64.55 | 216.24±60.81 | 0.332 |
| Shannon Index | 3.25±0.53 | 2.86±0.33 | 0.01 |
| OUT rate | 0.999 | 0.999 | 0.243 |
We performed Principal coordinates analysis (PCoA) analysis for Beta diversity analysis between the MM group and the healthy control group to detect the difference in species diversity among different samples (Figure 1). PCoA is a dimensionality reduction method, where each sample is shown as 1 point, and the square of the distance between points is equal to the original differential data, and the qualitative data is quantitatively converted. PCoA analysis ensures the sample classification. Briefly, closeness of samples on the graph indicates greater similarity. According to the PCoA analysis based on the Unifrac algorithm, the MM group and the control group were clustered separately, with good differentiation.
Figure 1.
Beta diversity of the gut microbiota in each group using PCoA analysis.
Species classification
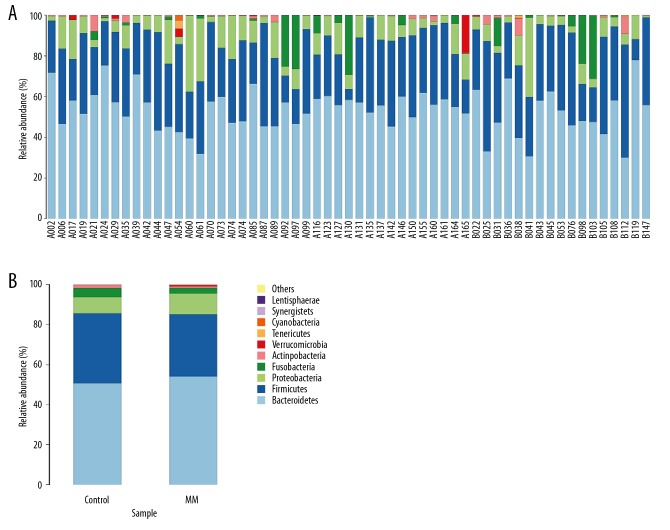
The representative sequence of OTU was compared with the microbial reference database to obtain species classification information corresponding to each OTU. The histogram of the species distribution of each sample at the phylum level is shown in Figure 2. In Figure 2, only 10 species with top abundance level are shown, while other species were combined into others and Unclassified represented species that have not been taxonomically annotated. Among the MM group and the healthy control group, Bacteroidetes, Firmicutes, Proteobacteria, Fusobacteria, Actinobacteria, Verrucomicrobia, and Teneriquetes were highly abundant. Bacteroidetes and Firmicutes were considered as the absolute dominant bacteria of IM, and the sequences of the 2 accounted for more than 80% of the total number of sequences.
Figure 2.
(A) Histogram of species distribution for each sample at the phylum level. The sample with the beginning of A indicates the MM group; the sample with the beginning of B indicates the control group. (B) Comparison of IM between the MM group and healthy control at the phylum level.
There was a significant difference in Proteobacteria and Actinomycetes in the IM between the MM group and healthy control group at the phylum level. Compared with the healthy control group, the proportion of Proteobacteria in the MM group was significantly increased (10.43% vs. 6.41%, p=0.021), and the proportion of Actinomycetes was significantly decreased (0.48% vs. 1.83%, p=0.035). There were no statistically significant differences in the other types of IM (Table 6).
Table 6.
Phylum-level comparison of IM between MM patients and healthy controls.
| Target flora phylum | Healthy controls (%, n=17) | MM (%, n=40) | P value |
|---|---|---|---|
| Bacteroidetes | 51.09±13.57 | 53.41±6.76 | 0.472 |
| Firmicutes | 35.13±13.83 | 31.51±10.49 | 0.358 |
| Proteobacteria | 6.41±3.91 | 10.43±5.52 | 0.021 |
| Fusobacteria | 5.31±10.20 | 3.99±8.73 | 0.669 |
| Actinobacteria | 1.83±3.02 | 0.48±0.75 | 0.035 |
| Verrucomicrobia | 0.06±1.74 | 0.16±0.52 | 0.492 |
| Tenericutes | 0.15±0.39 | 0.00±0.00 | 0.063 |
Differential significance analysis between groups
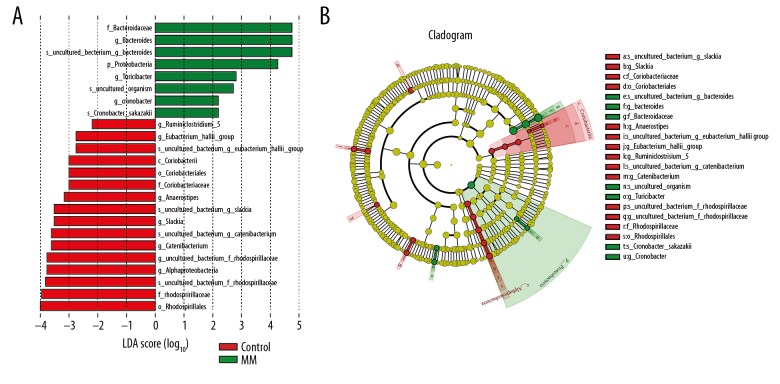
Differential significance analysis between groups was mainly designed to find the genus of bacteria between groups. We used LEfSE and Metastats to analyze the differential significance between groups. Line Discriminant Analysis Effect Size (LEfSE) showed significantly different bacteria between different groups. In this study, an LDA value of >2 was used as the cut-off value, and genera with LDA value greater than 2 are shown in Figure 3.
Figure 3.
Taxonomic differences were detected based on OTUs between MM group and healthy controls. (A) Linear discriminative analysis (LDA) effect size (LEfSe) analysis between healthy controls and MM patients. (B) Cladogram showing differentially abundant taxonomic clades with an LDA score >2.0 among patients and controls.
Metastats software was used to analyze taxonomic differences between different groups. The t test was performed on the species abundance data between the groups to obtain the P value, followed by final selection of the differential genus between 2 groups. The genus with significant difference between MM group and healthy control group was selected, followed by ordering according to the abundance in the intestinal tract of MM patients (as shown in Table 7). As a result, the intestinal level of Bacteroides, Faecalibacterium, and Roseburia of MM patients was significantly elevated. In addition, Anaeroplasma, Allobaculum, and Butyrivibrio were found to exist only in the MM group.
Table 7.
Differential analysis on genus between MM and healthy controls.
| Genus | Healthy control (n=17) | MM (n=40) | P value | ||
|---|---|---|---|---|---|
| Mean | SD | Mean | SD | ||
| Bacteroides | 3.10E-01 | 4.67E-02 | 4.17E-01 | 2.05E-02 | 0.046 |
| Faecalibacterium | 3.10E-02 | 6.29E-03 | 5.72E-02 | 9.40E-03 | 0.029 |
| Roseburia | 1.09E-02 | 3.69E-03 | 2.41E-02 | 5.25E-03 | 0.047 |
| Veillonella | 5.74E-04 | 2.09E-04 | 4.37E-03 | 2.61E-03 | 0.084 |
| Peptoclostridium | 3.59E-03 | 1.10E-03 | 1.40E-03 | 4.00E-04 | 0.09 |
| Acidaminococcus | 6.25E-05 | 3.98E-05 | 1.36E-03 | 8.40E-04 | 0.037 |
| Anaeroplasma | 0.00E+00 | 0.00E+00 | 1.03E-03 | 1.03E-03 | <0.001 |
| Dielma | 6.68E-06 | 3.96E-06 | 3.47E-04 | 3.06E-04 | 0.029 |
| Allobaculum | 0.00E+00 | 0.00E+00 | 4.09E-05 | 4.02E-05 | 0.097 |
| Nicotiana_otophora | 1.00E-05 | 4.36E-06 | 3.64E-05 | 1.29E-05 | 0.075 |
| Butyrivibrio | 0.00E+00 | 0.00E+00 | 2.27E-05 | 2.27E-05 | <0.001 |
Matching study in MM patients
The MM patients were matched with their family members in this study. A total of 21 pairs were obtained, and paired t test analysis was performed. As a result, the intestinal levels of Clostridium leptum and Pseudomonas aeruginosa were significantly higher in MM patients than in the healthy controls (P<0.05) (Table 8).
Table 8.
IM detection in MM patients and their family members using qRT-PCR.
| Target flora | Healthy controls (n=21) | MM (n=21) | P value |
|---|---|---|---|
| Enterococcus faecalis | 1.89±1.64 | 1.33±1.25 | 0.331 |
| Lactobacillus group | 2.86±0.82 | 3.30±1.27 | 0.178 |
| Clostridium cluster XI | 2.54±1.52 | 2.84±1.16 | 0.507 |
| Faecalibacterium prausnitzii | 4.49±1.52 | 4.98±1.57 | 0.032 |
| Pseudomonas alcaligenes | −0.17±1.21 | 0.36±1.44 | 0.334 |
| Pseudomonas aeruginosa | 1.49±1.36 | 2.77±1.37 | 0.043 |
| Gram-positive bacteria | 6.07±1.00 | 5.84±2.07 | 0.731 |
| Gram-negative bacteria | 4.03±1.18 | 4.23±1.87 | 0.628 |
Table 10.
MM comparison with metabolic genes in healthy controls.
| Class 1 | Class 2 | MM (%) | Healthy controls (%) | P value |
|---|---|---|---|---|
| Environmental information processing | Signal transduction | 2.85±0.35 | 2.50±0.17 | 0.0054 |
| Metabolism | Metabolism of terpenoids and polyketides | 1.86±0.06 | 1.94±0.05 | 0.0072 |
| Cellular processes | Cell growth and death | 0.67±0.06 | 0.73±0.05 | 0.0423 |
Correlation between Clostridium leptum and prognostic risk factors in MM patients
The levels of Clostridium leptum were compared in MM patients (N=41) with different ISS stages, extramedullary infiltration, LDH levels, and cytogenetic groups. The cut-off value of LDH level was 232 U/L, which was previously used in our department. FISH assay was used to detect genetic abnormalities such as 13q-, 17p-, 1q21, t(11;14) and t(4;14). Patients with high ISS stage (stage III) were found to have elevated levels of Clostridium leptum (Table 9). However, the high ISS stage was also affected by creatinine level (p=0.019) (as shown in Table 7). Meanwhile, Pearson correlation analysis indicated that the level of Clostridium leptum was significantly correlated with ISS stage (R=0.138, P=0.017), but the level of Clostridium leptum was not significantly associated with creatinine levels (P=0.170) (Figure 4).
Table 9.
The Clostridium leptum levels in MM patients with different prognostic risk factors.
| Risk factors | Expression | Clostridium leptum levels | P values |
|---|---|---|---|
| (Log 10 of copies/0.4g) | |||
| ISS stage | I–II (21) | 4.80±1.11 | 0.025 |
| III (19) | 5.49±0.76 | ||
| Extramedullary infiltration | +16 | 5.05±1.05 | 0.704 |
| −24 | 5.17±1.01 | ||
| LDH level | >232U/L (13) | 5.07±1.23 | 0.835 |
| <232U/L (27) | 5.15±0.93 | ||
| 13q- | +14 | 5.45±0.92 | 0.084 |
| −24 | 4.83±1.08 | ||
| 17p- | +6 | 4.97±1.04 | 0.784 |
| −32 | 5.10±1.06 | ||
| 1q21 | +15 | 5.28±0.71 | 0.347 |
| −23 | 4.95±1.21 | ||
| t (11;14) | 0 | – | - |
| −38 | 5.08±1.04 | ||
| t (4;14) | +8 | 5.51±0.74 | 0.184 |
| −30 | 4.96±1.09 |
Figure 4.
(A, B) Correlation analysis between the level of Clostridium leptum with ISS stage and creatinine levels.
Pathway changing
The sequences obtained from high-throughput sequencing were compared with the species composition information obtained from the 16S sequencing data using PICRUSt software. We therefore speculated on the composition of functional gene and analyzed the functional differences between the different groups.
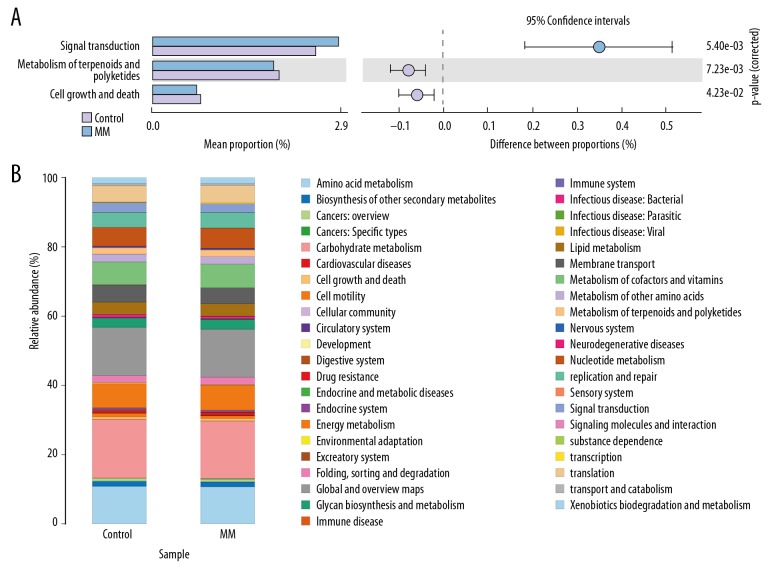
Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic pathway analysis was used for differential prediction and comparison of functional genes related to microbial community metabolic pathways between different samples. Compared with the control group, we found a significant increase in signal transduction gene expression (P=0.0054), whereas the gene of terpenoids and polyketides metabolism and cell proliferation and apoptosis genes were significantly decreased (P=0.0072, P=0.0423) (Table10, Figure 5).
Figure 5.
(A, B) Differential analysis of KEGG metabolic pathway between MM and healthy controls.
Discussion
Although previous studies have assessed the relationship between the intestinal microbiota and hematologic system, such as hematopoietic stem cell transplantation [21], bone marrow transplantation [22], lymphoid cells [23], and acute lymphoblastic leukemia [24], the association between intestinal microbiota and MM has rarely been investigated. In this study, 16s rDNA high-throughput sequencing was used to detect and analyze the difference in IM of MM patients, followed by relevant functional prediction. We found that Alpha diversity of IM was decreased in the MM group, suggesting the dysregulated IM in MM patients. In addition, Beta diversity analysis indicated differences in IM structure between MM patients and healthy controls. Phylum-level studies revealed that the proportion of Proteobacteria was higher in the MM group, while the proportion of the Actinobacteria was decreased. Genus-level studies showed that the levels of Bacteroides, Faecalibacterium, and Roseburia were increased. Species-level studies indicated that the proportion of E. coli, Bacteroides fragilis, and Pseudomonas aeruginosa was increased, while the proportion of Clostridium IX was decreased. Meanwhile, the increased level of Clostridium leptum was associated with ISS stage. Actinobacteria, Clostridium leptum, and Rothia are the main bacteria involved in the intestinal glucose metabolism pathway, suggesting the dysregulated glucose metabolism pathway in the intestinal tract of MM patients. Further KEGG metabolic functional gene predictive analysis revealed that the proportion of functional genes involved in the signal transduction pathway was higher in the MM group, and the proportion of functional genes involved in the metabolism of terpene and polyketides and apoptosis was lower.
Intestinal micro-ecology is a small dynamic ecological ecosystem composed of microorganisms and human beings, involved in the metabolism and immune processes of the human body. Recent studies have shown that the imbalance of intestinal micro-ecology not only can lead to gastrointestinal diseases, but also is associated with obesity, various types of digestive diseases, and multiple kinds of tumors [25,26]. The intestine, liver, and brain form the “gut-liver axis” and the “gut-brain axis”, which interact functionally [27,28]. In terms of research on the relationship between blood diseases and IM, most studies have focused on the investigation of IM changes involving bone marrow transplantation and chemotherapy agents [22,29,30], however, the relationship between MM and IM has been rarely investigated. In the present study, we used diversity analysis and microbiota analysis to investigate differences in IM structure in MM patients. Alpha diversity analysis showed that the species uniformity, Shannon index, was decreased, indicating decreased diversity of species uniformity and the dysregulation of IM in MM patients. IM diversity is an important indicator for maintaining homeostasis of the intestinal microenvironment. The decreased microbial diversity suggests the presence of dysregulation of the micro-ecology in MM patients. In Beta diversity analysis, PCoA principal coordinate analysis was used to explore the difference between MM patients and healthy controls. The PCoA principal coordinate analysis showed that the samples of MM patients and healthy controls were separately clustered, with good differentiation. The principal component analysis chart did not show any differential trend of the MMs of different heavy-chain types and light-chain types, indicating a significant difference of IM between MM patients and healthy controls. Although MM is a heterogeneous disease, there is no significant difference in IM structure between different types of MM patients.
We further analyzed the IM changes at the phylum, genus, and species levels. The phylum-level study of IM showed that Bacteroidetes, Firmicutes, Proteobacteria, Clostridium, Actinomyces, Verrucomicrobia, and Tenericutes had relatively higher abundance in the intestinal tract of the MM group and in the healthy control group. The intestinal proportion of Proteobacteria was increased and the intestinal proportion of Actinomyces was decreased in MM patients. Escherichia coli is the representative bacteria of Proteobacteria, which is the main bacteria in the intestine, and also is the infectious source of patients with hematological diseases during neutrophil-deficiency phase [31,32].
The genus-level study found that the levels of Bacteroides, Clostridium leptum, and Rothia were increased. Further studies revealed that the proportion of Bacteroides fragilis of Bacteroides, Clostridium leptum of Clostridium, Escherichia coli, and Pseudomonas aeruginosa were increased, while the proportion of Clostridium IX was decreased. Bacterial infection is one of the common complications of patients with blood diseases, and it is also one of the main causes of death, especially after chemotherapy. After chemotherapy, the level of neutrophils is reduced in the blood of patients, who become immunosuppressive hosts and are vulnerable to be infected, further progressing into septic shock with extremely high mortality [33]. It has been proved that the common infection sites of neutrophil deficiency-related infection are the respiratory tract, perianal, and gingival [34,35]. Respiratory infections are mainly due to Gram-positive bacteria, and perianal infections are mainly due to Gram-negative bacteria and anaerobic bacteria. In addition, an epidemiological survey showed that, among the Gram-negative bacterial infection, Enterobacteria represented by Escherichia coli and non-fermentative bacteria represented by Pseudomonas maltophilia and Pseudomonas aeruginosa accounted for the higher incidence of nosocomial infection [36,37]. In our study, we found that the intestinal abundance of Escherichia coli and Pseudomonas aeruginosa was increased in newly diagnosed MM patients, which was closely and consistently related to the pathogenic species causing bacterial infection in MM patients in clinical practice.
Our study found for the first time that the intestinal abundance of Actinobacteria was decreased in MM patients, while that of Clostridium and Rathio was increased. Correlation analysis of risk factors found that elevated levels of Clostridium leptum were associated with ISS stage but not creatinine levels. The main product of Actinobacteria is oligosaccharides, α-glucosidase inhibitors that inhibit the activity of various α-glucosidases such as maltase, isomaltase, glucoamylase, and invertase, thereby causing slower decomposing speed into glucose from starch and saccharose, thereby resulting in slow absorption of glucose in the intestine. Therefore, Actinobacteria is involved in glycometabolism regulation and participates in the decomposition from polysaccharides to glucose and fructose [38–40]. At present, the product application of Actinobacteria has been commercialized, which has been commercially applied in anti-diabetes agents, such as acarbose and voglibose.
Clostridium leptum and Rothia both belong to the Clostridialclusters family, but belong to different subgroups. Clostridium leptum belongs to subgroup IV, and Rothia belongs to subgroup XIVa [41]. Both of them are the main butyric acid-producing bacteria in the intestinal microenvironment. In the intestinal microenvironment, butyric acid is mainly produced by the metabolism of lactic acid, short-chain fatty acids, and acetic acid in the sugar metabolism pathway. Butyric acid is the main energy source of intestinal mucosal cells, and also accounts for approximately 10% of the human body’s energy source. Both Clostridium leptum and Rothia are involved in the process of acetic acid metabolism to produce butyric acid through butyryl-CoA: acetyl-CoA transferase [41], which is involved in glucose regulation from glucose to butyric acid.
The bacterial products of Actinobacteria are involved in the decomposition regulation of polysaccharides into monosaccharides in the sugar metabolism pathway. Butyric acid-producing bacteria, including Clostridium leptum and Rothia, are involved in the butyric acid production from glucose metabolism in the sugar metabolism pathway. The changes of intestinal Actinobacteria, Clostridium leptum, and Rothia in MM patients suggest that MM patients have dysregulation of the sugar metabolism pathway in the intestine compared with the healthy controls. The decreased proportion of Actinobacteria suggests that the dysregulation of the glucose metabolism pathway in MM patients is mainly manifested as the increased conversion from starch and other polysaccharides into glucose. The increased proportion of butyric acid-producing bacteria, such as Clostridium leptum and Rothia, suggests that the dysregulation of the glucose metabolism pathway in MM patients is mainly manifested as the increased conversion from glucose into butyrate. However, animal experiments and metabolomics experiments are needed to further clarify the specific mechanism underlying the relationship between the dysregulated glucose metabolism pathway and MM.
Further differential analysis of the KEGG metabolic pathway revealed that the level of functional genes involved in signal transduction was increased in MM group compared to healthy controls, while the levels of functional genes involved in the metabolism of terpene and polyketides and apoptosis was decreased, suggesting that the functional gene changes in the signaling transduction and metabolism in IM in MM patients. We suppose that it may due to some stress responses of the body system against the disease, but this needs further research to validate. Terpene and polyketides are metabolites of bacteria and fungi, which are produced by continuous condensation reactions from lower carboxylic acid [42]. Polyketides are mainly involved in intercellular signal transduction processes, including macrolides, tetracyclines, and anthracyclines. Among them, type I polyketide synthase is used in macrolides, and type II polyketide synthase is involved in the synthesis of tetracyclines and anthracyclines [43–45].
Conclusions
We performed a large-scale investigation of the changes of the main IM in MM patients. We analyzed the IM changes at phylum, genus, and species levels, validated the IM between MM patients, and matched relatives using RT-PCR, which revealed the increased intestinal levels of Clostridium leptum and Pseudomonas aeruginosa in MM patients. Further correlation analysis show that the elevated level of Clostridium leptum is related to the ISS stage, providing new ideas and potential targets for the diagnosis and therapy of MM. Further studies will focus on the in vitro and in vivo validation of these results and assess the regulation mechanism of the glucose metabolism pathways.
Footnotes
Conflict of interests
None.
Source of support: Departmental sources
References
- 1.Sidhu G, Homsi Y. Severe lytic bone lesions in multiple myeloma. Am J Med Sci. 2019;357:e11–12. doi: 10.1016/j.amjms.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 2.Alexander DD, Mink PJ, Adami HO, et al. Multiple myeloma: A review of the epidemiologic literature. Int J Cancer. 2007;120:40–61. doi: 10.1002/ijc.22718. [DOI] [PubMed] [Google Scholar]
- 3.Lindqvist EK, Goldin LR, Landgren O, et al. Personal and family history of immune-related conditions increase the risk of plasma cell disorders: A population-based study. Blood. 2011;118:6284–91. doi: 10.1182/blood-2011-04-347559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weinstock GM. Genomic approaches to studying the human microbiota. Nature. 2012;489:250. doi: 10.1038/nature11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flint HJ, Bayer EA, Rincon MT, et al. Polysaccharide utilization by gut bacteria: Potential for new insights from genomic analysis. Nat Rev Microbiol. 2008;6:121–31. doi: 10.1038/nrmicro1817. [DOI] [PubMed] [Google Scholar]
- 6.Flint HJ, Scott KP, Louis P, Duncan SH. The role of the gut microbiota in nutrition and health. Nat Rev Gastroenterol Hepatol. 2012;9:577–89. doi: 10.1038/nrgastro.2012.156. [DOI] [PubMed] [Google Scholar]
- 7.Manco M, Putignani L, Bottazzo GF. Gut microbiota, lipopolysaccharides, and innate immunity in the pathogenesis of obesity and cardiovascular risk. Endocr Rev. 2010;31:817–44. doi: 10.1210/er.2009-0030. [DOI] [PubMed] [Google Scholar]
- 8.Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. 2016;535:75–84. doi: 10.1038/nature18848. [DOI] [PubMed] [Google Scholar]
- 9.Gilbert JA, Quinn RA, Debelius J, et al. Microbiome-wide association studies link dynamic microbial consortia to disease. Nature. 2016;535:94–103. doi: 10.1038/nature18850. [DOI] [PubMed] [Google Scholar]
- 10.Kostic AD, Chun E, Robertson L, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–15. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boleij A, Hechenbleikner EM, Goodwin AC, et al. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis. 2014;60:208–15. doi: 10.1093/cid/ciu787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157:121–41. doi: 10.1016/j.cell.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Avilés-Jiménez F, Guitron A, Segura-López F, et al. Microbiota studies in the bile duct strongly suggest a role for Helicobacter pylori in extrahepatic cholangiocarcinoma. Clin Microbiol Infect. 2016;22:178.e11–e22. doi: 10.1016/j.cmi.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 14.O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep. 2006;7:688–93. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lakritz JR, Poutahidis T, Mirabal S, et al. Gut bacteria require neutrophils to promote mammary tumorigenesis. Oncotarget. 2015;6:9387–96. doi: 10.18632/oncotarget.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fox JG, Feng Y, Theve EJ, et al. Gut microbes define liver cancer risk in mice exposed to chemical and viral transgenic hepatocarcinogens. Gut. 2010;59:88–97. doi: 10.1136/gut.2009.183749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Poutahidis T, Cappelle K, Levkovich T, et al. Pathogenic intestinal bacteria enhance prostate cancer development via systemic activation of immune cells in mice. PLoS One. 2013;8:e73933. doi: 10.1371/journal.pone.0073933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rajagopala SV, Yooseph S, Harkins DM, et al. Gastrointestinal microbial populations can distinguish pediatric and adolescent Acute Lymphoblastic Leukemia (ALL) at the time of disease diagnosis. BMC Genomics. 2016;17:635. doi: 10.1186/s12864-016-2965-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greipp PR, San Miguel J, Durie BG, et al. International staging system for multiple myeloma. J Clin Oncol. 2005;23:3412–20. doi: 10.1200/JCO.2005.04.242. [DOI] [PubMed] [Google Scholar]
- 20.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 21.Taur Y, Jenq RR, Perales M-A, et al. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood. 2014;124:1174–82. doi: 10.1182/blood-2014-02-554725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peled JU, Jenq RR, Holler E, et al. Role of gut flora after bone marrow transplantation. Nat Microbiol. 2016;1:16036. doi: 10.1038/nmicrobiol.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pearson C, Uhlig HH, Powrie F. Lymphoid microenvironments and innate lymphoid cells in the gut. Trends Immunol. 2012;33:289–96. doi: 10.1016/j.it.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 24.Rajagopala SV, Yooseph S, Harkins DM, et al. Gastrointestinal microbial populations can distinguish pediatric and adolescent Acute Lymphoblastic Leukemia (ALL) at the time of disease diagnosis. BMC Genomics. 2016;17:635. doi: 10.1186/s12864-016-2965-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sekirov I, Russell SL, Antunes LCM, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- 26.Nicholson JK, Holmes E, Kinross J, et al. Host-gut microbiota metabolic interactions. Science. 2012;336:1262–67. doi: 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- 27.Mayer EA, Tillisch K, Gupta A. Gut/brain axis and the microbiota. J Clin Invest. 2015;125:926–38. doi: 10.1172/JCI76304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Compare D, Coccoli P, Rocco A, et al. Gut–liver axis: The impact of gut microbiota on non alcoholic fatty liver disease. Nutr Metab Cardiovasc Dis. 2012;22:471–76. doi: 10.1016/j.numecd.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 29.Montassier E, Batard E, Massart S, et al. 16S rRNA gene pyrosequencing reveals shift in patient faecal microbiota during high-dose chemotherapy as conditioning regimen for bone marrow transplantation. Microb Ecol. 2014;67:690–99. doi: 10.1007/s00248-013-0355-4. [DOI] [PubMed] [Google Scholar]
- 30.Vossen JM, Guiot HF, Lankester AC, et al. Complete suppression of the gut microbiome prevents acute graft-versus-host disease following allogeneic bone marrow transplantation. PLoS One. 2014;9:e105706. doi: 10.1371/journal.pone.0105706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ben-Chetrit E, Eldaim MA, Bar-Meir M, et al. Associated factors and clinical outcomes of bloodstream infection due to extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae during febrile neutropenia. Int J Antimicrob Agents. 2019;53:423–28. doi: 10.1016/j.ijantimicag.2018.12.003. [DOI] [PubMed] [Google Scholar]
- 32.Viscoli C, Varnier O, Machetti M. Infections in patients with febrile neutropenia: epidemiology, microbiology, and risk stratification. Clin Infect Dis. 2005;40(Suppl 4):S240–45. doi: 10.1086/427329. [DOI] [PubMed] [Google Scholar]
- 33.Hotchkiss RS, Karl IE. The pathophysiology and treatment of sepsis. N Engl J Med. 2003;348:138–50. doi: 10.1056/NEJMra021333. [DOI] [PubMed] [Google Scholar]
- 34.Mizgerd JP. Acute lower respiratory tract infection. N Engl J Med. 2008;358:716–27. doi: 10.1056/NEJMra074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lakshman R, Finn A. Neutrophil disorders and their management. J Clin Pathol. 2001;54:7–19. doi: 10.1136/jcp.54.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaynes R1, Edwards JR National Nosocomial Infections Surveillance System. Overview of nosocomial infections caused by gram-negative bacilli. Clin Infect Dis. 2005;41:848–54. doi: 10.1086/432803. [DOI] [PubMed] [Google Scholar]
- 37.Khan HA, Ahmad A, Mehboob R. Nosocomial infections and their control strategies. Asian Pac J Trop Biomed. 2015;5:509–14. [Google Scholar]
- 38.Hu J-L, Nie S-P, Xie M-Y. Antidiabetic mechanism of dietary polysaccharides based on their gastrointestinal functions. J Agric Food Chem. 2018;66:4781–86. doi: 10.1021/acs.jafc.7b05410. [DOI] [PubMed] [Google Scholar]
- 39.Fang Q, Hu J, Nie Q, Nie S. Effects of polysaccharides on glycometabolism based on gut microbiota alteration. Trends in Food Science & Technology. 2019;92:65–70. [Google Scholar]
- 40.Binda C, Lopetuso LR, Rizzatti G, et al. Actinobacteria: A relevant minority for the maintenance of gut homeostasis. Dig Liver Dis. 2018;50:421–28. doi: 10.1016/j.dld.2018.02.012. [DOI] [PubMed] [Google Scholar]
- 41.Louis P, Flint HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. 2009;294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x. [DOI] [PubMed] [Google Scholar]
- 42.Mousa WK, Athar B, Merwin NJ, Magarvey NA. Antibiotics and specialized metabolites from the human microbiota. Nat Prod Rep. 2017;34:1302–31. doi: 10.1039/c7np00021a. [DOI] [PubMed] [Google Scholar]
- 43.Moloney MG. Natural products as a source for novel antibiotics. Trends Pharmacol Sci. 2016;37:689–701. doi: 10.1016/j.tips.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 44.Yuzawa S, Keasling JD, Katz L. Insights into polyketide biosynthesis gained from repurposing antibiotic-producing polyketide synthases to produce fuels and chemicals. J Antibiot. 2016;69:494–99. doi: 10.1038/ja.2016.64. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Z, Pan H-X, Tang G-L. New insights into bacterial type II polyketide biosynthesis. F1000Res. 2017;6:172. doi: 10.12688/f1000research.10466.1. [DOI] [PMC free article] [PubMed] [Google Scholar]