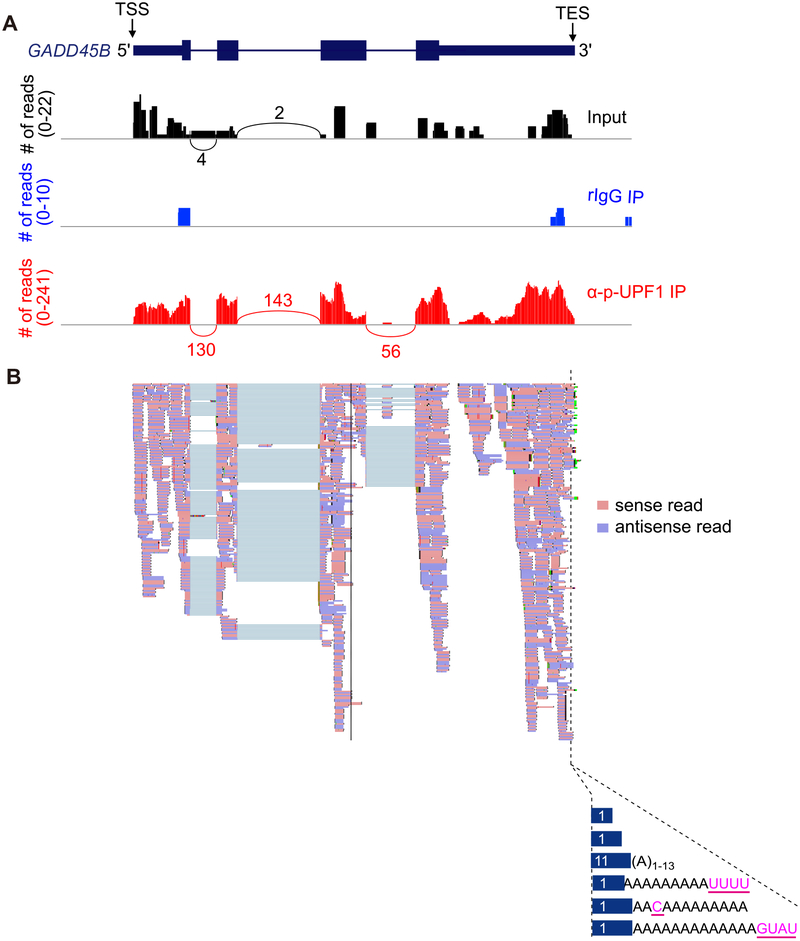
Fig. 3.
Results from NMD-DegSeq demonstrate the presence of nontemplated nucleotide additions at the 3′ends of p-UFP1-bound NMD decay intermediates. (A) Sashimi plots (black, Input; blue, rIgG IP; red, anti-p-UPF1 IP) developed using Integrative Genomics Viewer software [24] exemplifying NMD-DegSeq-read mapping to growth arrest and DNA-damage-inducible beta (GADD45B) mRNA, which is a bona fide NMD target, superimposed on the GADD45B gene as shown in the uppermost diagram [5,11]. TSS, transcript start site; TES, mature transcript end site, i.e. the site to which poly(A) is appended; numbers above or below arcs denote read numbers. (B) Using data shown in A for the p-UFP1 IP sample, individual sense reads (pink) and antisense reads (light purple) are shown (upper) and aligned with nontemplated nucleotide additions (pink and underlined; lower) observed within the poly(A) tail of GADD45B transcripts. Numbers in blue bars specify the number of paired reads having the corresponding 3′-end.