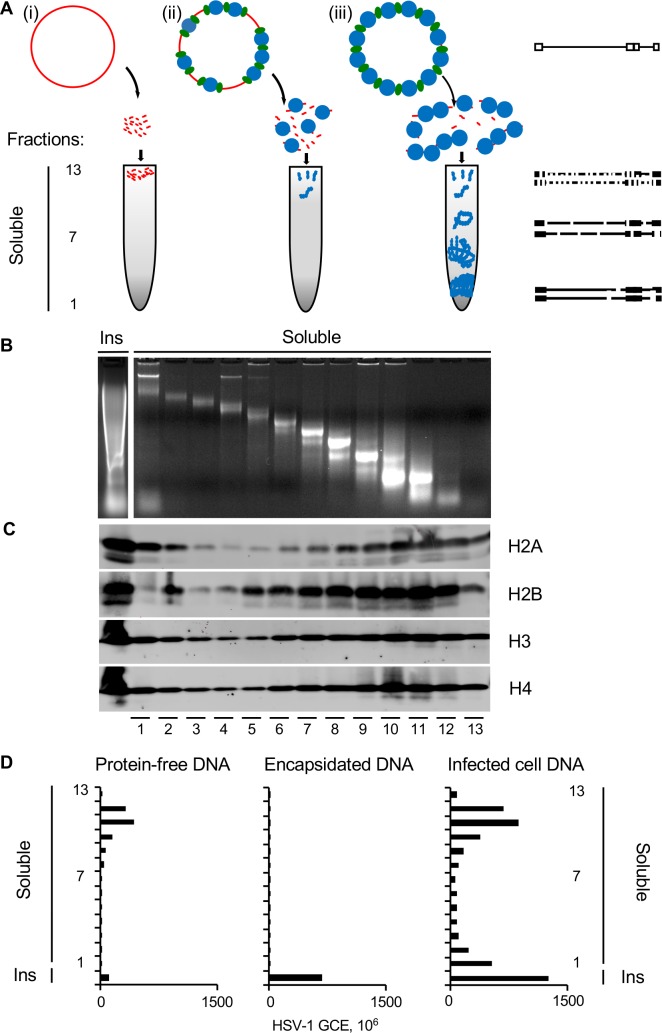
Fig 2. Intracellular HSV-1 DNA is differentially protected from MCN digestion than deproteinized or encapsidated DNA.
(A) Cartoon presenting the different models proposed for the HSV-1 chromatin and the expected fractionation patterns after serial digestion and sucrose gradients of: (i) mostly non-chromatinized HSV-1 DNA, (ii) HSV-1 DNA associated with nucleosomes only sporadically, (iii) fully (stably or unstably) chromatinized HSV-1 DNA. (B) DNA from each fraction was resolved in 2% agarose gel electrophoresis and stained with EtBr. (C) Western blot for core histones (H2A, H2B, H3 and H4) in all fractions, each resolved by SDS-PAGE and probed with corresponding antibodies. (D) Bar graph showing the number of HSV-1 genome copy equivalents in each fraction. Chromatin of HSV-1 infected cells was digested and fractionated. DNA in each fraction was subjected to deep sequencing. Ins, insoluble chromatin; GCE, genome copy equivalents. Results from one experiment representative of three.