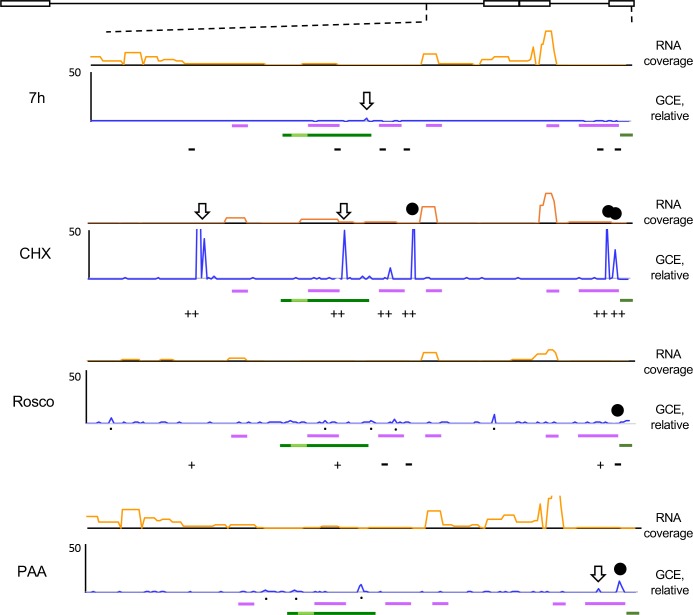
Fig 10. Enlargement of the repeats region, from position 100kbp to 152kbp, of the plots presented in Fig 9.
Orange lines graphs, HSV-1 RNA reads. X-axes, genome position (cartoon on top). Blue lines, HSV-1 number of genome copy equivalents (GCE) in each genome position. Black downward empty arrows, CTCF binding sites in strain 17; black solid circles, chromatin insulator-like elements; black dots, other overrepresented short sequences; purple bars underneath genome plots, IE genes; dark green bars underneath genome plots, LAT; light green bar, stable LAT; GCE, genome copy equivalent. Results from one experiment representative of three. ++ sequences co-immunoprecipitated with CTCF by ChIP in two independent experiments; + sequences co-immunoprecipitated with CTCF by ChIP in one of two independent experiments;—sequences not co-immunoprecipitated with CTCF by ChIP in either of the two independent experiments.