Fig 8. Mutation of EGR1 binding sites blocks induction of UL138.
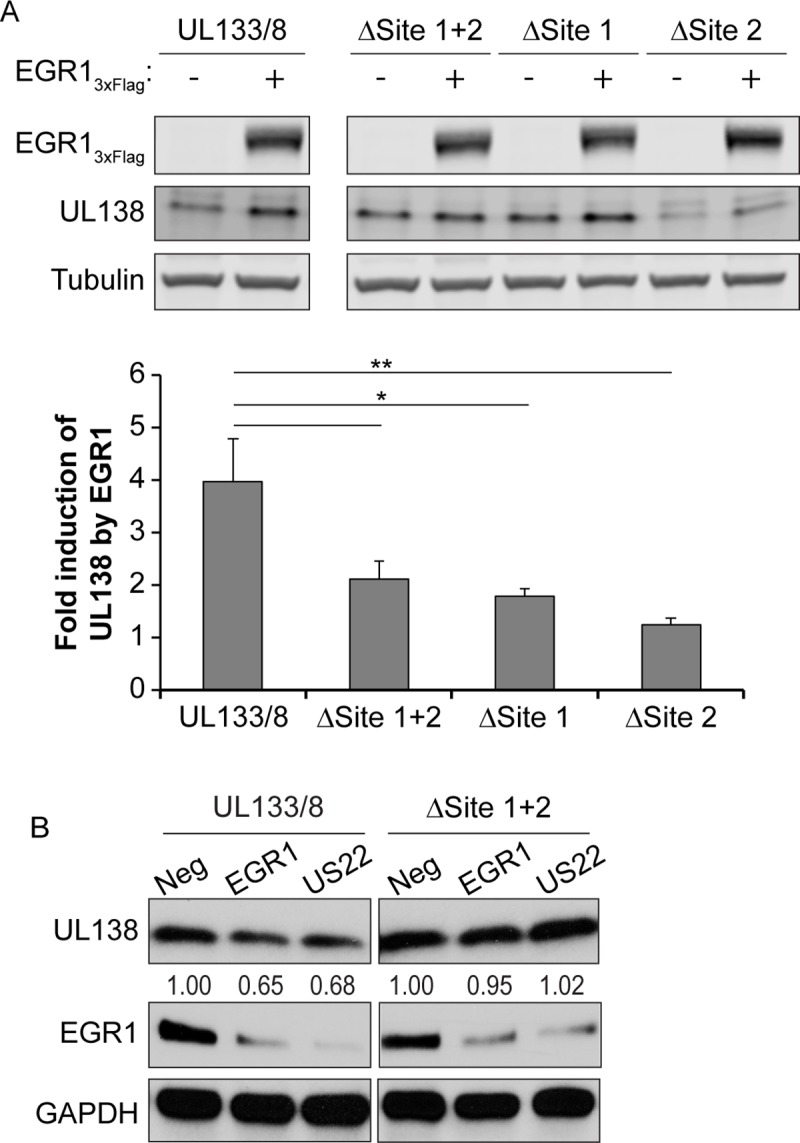
(A) HEK293T cells were co-transfected with either empty vector (minus sign) or EGR13xFLAG and a promoterless plasmid containing UL133/8 sequences or UL133/8 where EGR1 sites were mutated in combination (ΔSite1+2) or individually (ΔSite 1 or ΔSite 2). At 48 h, lysates were separated by SDS-PAGE, and proteins detected using α-Flag, α-UL138, and α-tubulin. UL138 protein levels in EGR13xFLAG transfections were normalized to control levels to determine UL138 induction. The results from 4 independent replicates are graphed. Statistical significance was calculated by One-Way ANOVA with Bonferroni correction (* p-value < 0.05 and ** p-value < 0.01). (B) HEK293T cells were co-transfected with the UL133/8 vector or the UL133/8 vector where EGR1 sites (ΔSite1, ΔSite 2) were disrupted and negative control siRNA, EGR1 siRNA, or miR-US22. Cells were transferred to serum-free media at 24 h. At 48 h, samples were stimulated with 50 ng/mL EGF for 1h and then lysed, separated by SDS-PAGE, and proteins detected using α-UL138 and α-GAPDH. UL138 levels are normalized to negative control. A representative blot of 2 independent experiments is shown.
