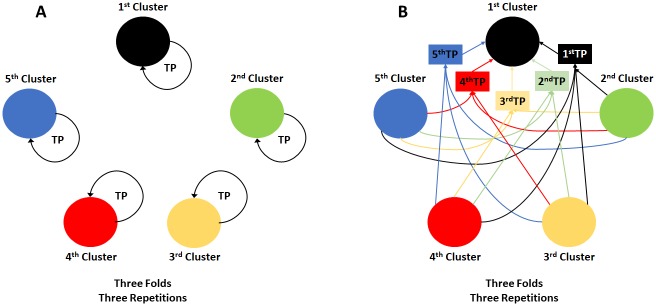
Fig 1. Cross validations performed to estimate the effect of population structure in genomic selection.
A. 2nd cross validation strategy–Within-cluster cross validation formed by discriminant analysis of principal components (DAPC); B. Illustration of between-clusters cross validation for DAPC cluster 1: 3rd cross-validation strategy–cross validation with training population composed of a third part of DAPC clusters 2–5 and validation in a third part of DAPC cluster 1 (1stTP); 4th cross-validation strategy–cross validation with the training population composed by a third part of three of the DAPC clusters (2nd, 3rd 4th, and 5th TPs). Therefore, the first training population (1st TP) is formed from the four remaining DAPC clusters (3rd cross validation strategy), while the second, third, fourth and fifth training populations (2nd, 3rd 4th and 5th TPs) were each created from only three of those four DAPC clusters (4th cross validation strategy).