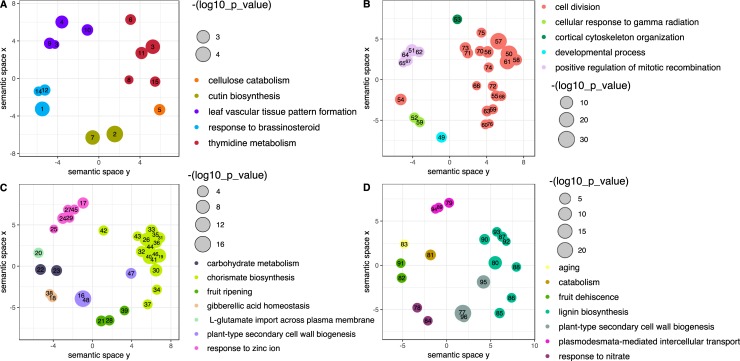
Fig 5. REVIGO scatter plots of enriched biological process GO terms for oak genes expressed during development of galled and normal oak bud tissues.
A: GO terms that were upregulated only in Early versus Growth stage gall tissues, and not in comparisons between any normal (ungalled) bud stages. B: GO terms that were upregulated both in Early versus Growth stage gall tissues and in normal buds (S4) versus open leaves (S5). C: GO terms that were downregulated only in Early versus Growth stage gall tissues, and not in comparisons between any normal (ungalled) bud stages. D: GO terms that were downregulated both in Early versus Growth stage gall tissues and in normal buds (S4) versus open leaves (S5). Circle size is scaled by–log10 of the enrichment p-value with scales inset next to each plot. Groups of GO terms are coloured by cluster as classified by REVIGO treemaps (S3 Fig). There are fewer GO terms than for the equivalent TopGO comparisons as REVIGO reduced the redundancy in enriched GO terms. Numbers in the figures refer to the following GO terms and reference numbers: 1 = response to brassinosteroid (GO:0009741), 2 = cutin biosynthetic process (GO:0010143), 3 = thymidine metabolic process (GO:0046104), 4 = leaf vascular tissue pattern formation (GO:0010305), 5 = cellulose catabolic process (GO:0030245), 6 = asymmetric cell division (GO:0008356), 7 = wax biosynthetic process (GO:0010025), 8 = lignan biosynthetic process (GO:0009807), 9 = regulation of organ growth (GO:0046620), 10 = guard cell differentiation (GO:0010052), 11 = fatty acid metabolic process (GO:0006631), 12 = auxin-activated signalling pathway (GO:0009734), 13 = regulation of seed germination (GO:0010029), 14 = cytokinin-activated signalling pathway (GO:0009736), 15 = lipid catabolic process (GO:0016042), 16 = plant-type secondary cell wall biogenesis (GO:0009834), 17 = response to zinc ion (GO:0010043), 18 = gibberellic acid homeostasis (GO:0010336), 19 = chorismate biosynthetic process (GO:0009423), 20 = L-glutamate import across plasma membrane (GO:0098712), 21 = fruit ripening (GO:0009835), 22 = carbohydrate metabolic process (GO:0005975), 23 = proteolysis (GO:0006508), 24 = response to osmotic stress (GO:0006970), 25 = wound healing (GO:0042060), 26 = oxidation-reduction process (GO:0055114), 27 = response to herbicide (GO:0009635), 28 = regeneration (GO:0031099), 29 = response to biotic stimulus (GO:0009607), 30 = lignin biosynthetic process (GO:0009809), 31 = one-carbon metabolic process (GO:0006730), 32 = S-adenosylhomocysteine catabolic process (GO:0019510), 33 = shikimate metabolic process (GO:0019632), 34 = UDP-N-acetylglucosamine biosynthetic process (GO:0006048), 35 = ether metabolic process (GO:0018904), 36 = nitrate assimilation (GO:0042128), 37 = S-adenosylmethionine biosynthetic process (GO:0006556), 38 = sodium ion homeostasis (GO:0055078), 39 = mucilage biosynthetic process involved in seed coat development (GO:0048354), 40 = aromatic amino acid family biosynthetic process (GO:0009073), 41 = gluconeogenesis (GO:0006094), 42 = ureide catabolic process (GO:0010136), 43 = uracil catabolic process (GO:0006212), 44 = oxidative phosphorylation (GO:0006119), 45 = response to water deprivation (GO:0009414), 46 = beta-alanine biosynthetic process (GO:0019483), 47 = glucuronoxylan biosynthetic process (GO:0010417), 48 = cell wall organization (GO:0071555), 49 = developmental process (GO:0032502), 50 = cell division (GO:0051301), 51 = positive regulation of mitotic recombination (GO:0045951), 52 = cellular response to gamma radiation (GO:0071480), 53 = cortical cytoskeleton organization (GO:0030865), 54 = cell proliferation (GO:0008283), 55 = guard mother cell differentiation (GO:0010444), 56 = cytokinesis by cell plate formation (GO:0000911), 57 = microtubule-based movement (GO:0007018), 58 = microtubule-based process (GO:0007017), 59 = thigmotropism (GO:0009652), 60 = stomatal complex patterning (GO:0010375), 61 = cell cycle (GO:0007049), 62 = regulation of transcription, DNA-templated (GO:0006355), 63 = cotyledon development (GO:0048825), 64 = positive regulation of ubiquitin protein ligase activity (GO:1904668), 65 = positive regulation of anthocyanin metabolic process (GO:0031539), 66 = maintenance of floral organ identity (GO:0048497), 67 = positive regulation of cell proliferation (GO:0008284), 68 = epidermal cell fate specification (GO:0009957), 69 = endosperm development (GO:0009960), 70 = anastral spindle assembly involved in male meiosis (GO:0009971), 71 = DNA endoreduplication (GO:0042023), 72 = trichome branching (GO:0010091), 73 = mitotic spindle assembly checkpoint (GO:0007094), 74 = meiotic cell cycle (GO:0051321), 75 = phragmoplast microtubule organization (GO:0080175), 76 = polarity specification of adaxial/abaxial axis (GO:0009944), 77 = plant-type secondary cell wall biogenesis (GO:0009834), 78 = response to nitrate (GO:0010167), 79 = plasmodesmata-mediated intercellular transport (GO:0010497), 80 = lignin biosynthetic process (GO:0009809), 81 = catabolic process (GO:0009056), 82 = fruit dehiscence (GO:0010047), 83 = aging (GO:0007568), 84 = cellular response to salt stress (GO:0071472), 85 = S-adenosylmethionine biosynthetic process (GO:0006556), 86 = ethylene biosynthetic process (GO:0009693), 87 = regulation of salicylic acid metabolic process (GO:0010337), 88 = 3'-UTR-mediated mRNA destabilization (GO:0061158), 89 = nitrate transport (GO:0015706), 90 = oxidation-reduction process (GO:0055114), 91 = fruit ripening (GO:0009835), 92 = glycine catabolic process (GO:0006546), 93 = nitrate assimilation (GO:0042128), 94 = oligopeptide transport (GO:0006857), 95 = glucuronoxylan biosynthetic process (GO:0010417), 96 = cell wall organization (GO:0071555).