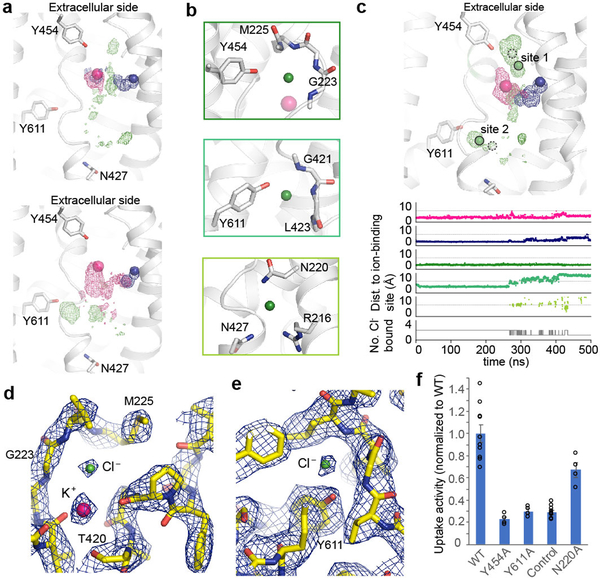
Figure 4 |. Chloride-binding sites.
a, Probability density for potassium (pink), sodium (blue), or chloride (green) across two simulations initiated with bound cations. Spheres indicate the initial position of cations. b, Three regions of chloride binding identified in simulation. c, Ion probability densities (top), as shown in a, for a simulation initiated with all four ions bound. The initial position of K+, Na+, and Cl− are indicated as pink, blue, and green (dashed boundaries) spheres, respectively. The positions of Cl− based on the cryo-EM are indicated by green spheres with solid boundaries. First five traces (bottom) show distance from ion-binding site to nearest ion of that type (see Methods). In the presence of stably bound chlorides, cations reside longer within the translocation pore. Light grey lines at 5 Å offer visual guide. Bottom trace shows number of chlorides occupying the chloride-binding sites. d, Upper chloride-binding site (site 1) and potassium-binding site density map (11.0 σ, blue mesh). e, Lower chloride-binding site (site 2) density map (6.0 σ, blue mesh). f, Mutant uptake activities, normalized to WT (mean±s.e.m., n=4 independent experiments; WT and control are the same as in Fig. 1e).