Abstract
Clavulanic acid is a bacterial specialized metabolite, which inhibits certain serine β-lactamases, enzymes that inactivate β-lactam antibiotics to confer resistance. Due to this activity, clavulanic acid is widely used in combination with penicillin and cephalosporin (β-lactam) antibiotics to treat infections caused by β-lactamase-producing bacteria. Clavulanic acid is industrially produced by fermenting Streptomyces clavuligerus, as large-scale chemical synthesis is not commercially feasible. Other than S. clavuligerus, Streptomyces jumonjinensis and Streptomyces katsurahamanus also produce clavulanic acid along with cephamycin C, but information regarding their genome sequences is not available. In addition, the Streptomyces contain many biosynthetic gene clusters thought to be “cryptic,” as the specialized metabolites produced by them are not known. Therefore, we sequenced the genomes of S. jumonjinensis and S. katsurahamanus, and examined their metabolomes using untargeted mass spectrometry along with S. clavuligerus for comparison. We analyzed the biosynthetic gene cluster content of the three species to correlate their biosynthetic capacities, by matching them with the specialized metabolites detected in the current study. It was recently reported that S. clavuligerus can produce the plant-associated metabolite naringenin, and we describe more examples of such specialized metabolites in extracts from the three Streptomyces species. Detailed comparisons of the biosynthetic gene clusters involved in clavulanic acid (and cephamycin C) production were also performed, and based on our analyses, we propose the core set of genes responsible for producing this medicinally important metabolite.
Keywords: Streptomyces, specialized metabolism, metabolomics, genomics, gene clusters, β-lactams, clavulanic acid
Introduction
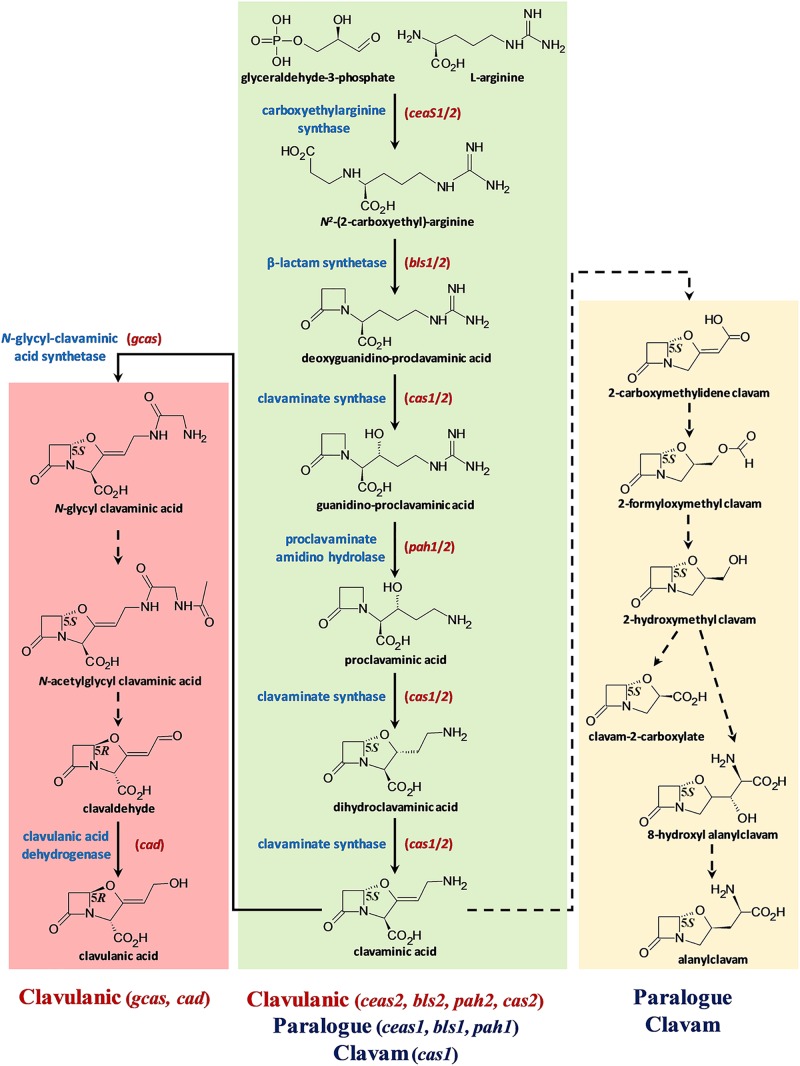
Bacteria from the genus Streptomyces produce numerous and diverse specialized (or secondary) metabolites (SMs), many of which have medicinal applications (Baltz, 2008). Some of these SMs are also used as antibiotic adjuvants, agents administered in conjunction with antibiotics to potentiate or restore their activities against resistant bacteria (Tyers and Wright, 2019). Clavulanic acid (CA, a 5R clavam SM, Figure 1) is an irreversible inhibitor of certain class A and D serine β-lactamases, which are enzymes that hydrolyze β-lactam antibiotics such as the penicillins and cephalosporins to confer resistance (Drawz and Bonomo, 2010). Therefore, CA is widely used in human and veterinary medicine in combination with β-lactam antibiotics to treat otherwise resistant infections caused by β-lactamase-producing bacteria (Brown, 1986).
FIGURE 1.
The S. clavuligerus clavulanic acid and 5S clavam biosynthetic pathways. The pathway is depicted in three parts: the central green box represents the steps shared between CA and 5S clavam biosynthesis, whereas the pink (left) and yellow (right) boxes indicate the late steps specific for CA or 5S clavam production, respectively. The solid arrows represent known reactions and broken arrows indicate uncharacterized steps, which could potentially involve more than one unknown gene product/enzyme. The names of core biosynthetic enzymes (blue) catalyzing known reactions and the respective gene(s) encoding them (red) are included where applicable. The stereochemistries (R/S) of the intermediates/products are also included along with their names. The identities of the gene clusters involved in each stage of biosynthesis is indicated at the bottom of the figure. Note that the shared part of the pathway (green) involves substitutable isozymes (CeaS, Bls, Cas, and Pah), which are encoded by two sets of genes (1 and 2) residing in three separate gene clusters. Additional genes from the respective clusters for which exact biosynthetic functions have not been assigned are not shown to simply interpretation.
Clavulanic acid is industrially produced by fermenting the bacterium Streptomyces clavuligerus (Jensen and Paradkar, 1999; Townsend, 2002; Saudagar et al., 2008), which was first identified during screens for microorganisms capable of producing β-lactam antibiotics such as cephamycin C (Ceph-C) (Brown et al., 1976). Apart from S. clavuligerus, Streptomyces jumonjinensis and Streptomyces katsurahamanus are the only other species known to produce CA along with Ceph-C (Ward and Hodgson, 1993; Jensen, 2012). In addition, CA production in S. clavuligerus generally occurs in conjunction with Ceph-C (Romero et al., 1984; Jensen and Paradkar, 1999), even though both metabolites are products of distinct biosynthetic pathways (Hamed et al., 2013). As in the case of other Actinobacterial SMs (van der Heul et al., 2018), the regulation of CA production in S. clavuligerus is complex and involves cluster-situated regulators, global mechanisms, and signaling cascades (Liras et al., 2008; Song et al., 2010a; Paradkar, 2013; Ferguson et al., 2016; Álvarez-Álvarez et al., 2017). S. clavuligerus is also unique among the CA producers described so far due to its ability to produce the structurally related 5S clavams (Brown et al., 1979; Pruess and Kellett, 1983), which partially share a common biosynthetic pathway with CA (Figure 1; Egan et al., 1997; Jensen, 2012). The 5S clavams have the opposite stereochemistry as compared to CA and are therefore not inhibitory toward β-lactamases, but instead some of them display weak antibacterial, antifungal, or antimetabolite activities (Jensen, 2012). In comparison, some Streptomyces species only synthesize the 5S clavams but not CA, suggesting that the ability to produce clavams with both stereochemistries (5R and 5S, Figure 1) might be unique to S. clavuligerus (Jensen and Paradkar, 1999; Challis and Hopwood, 2003).
It is now recognized that the Streptomyces contain many biosynthetic gene clusters (BGCs) thought to be “cryptic or silent,” as the SMs produced by them are not known (Katz and Baltz, 2016). On average, each Streptomyces species contains ∼30 BGCs but only produces 3–5 SMs under laboratory conditions. Additionally, recent reports have shown that S. clavuligerus produced some SMs only thought to originate from plants (Álvarez-Álvarez et al., 2015), highlighting the need for thoroughly cataloging specialized metabolism, even from well-studied organisms. Due to the small number of Streptomyces species known to produce CA, it is of interest to determine if S. clavuligerus, S. jumonjinensis, and S. katsurahamanus also share other metabolic capabilities. Therefore, we sequenced the genomes of S. jumonjinensis and S. katsurahamanus, conducted comparative metabolomics analysis on the three CA producers to identify SMs, and correlated their biosynthesis with predicted BGCs wherever possible.
The described analyses also provide information on BGC content from S. jumonjinensis and S. katsurahamanus, insight that was not available previously. In S. clavuligerus, three separate BGCs are involved in clavam metabolite biosynthesis (Tahlan et al., 2004a). The clavulanic acid BGC is primarily associated with CA production (Jensen et al., 2000, 2004a; Li et al., 2000; Mellado et al., 2002), whereas the clavam and paralog BGCs are involved in the biosynthesis of the 5S clavams (Figure 1; Tahlan et al., 2007; Zelyas et al., 2008). Because of the common biosynthetic origins of CA and the 5S clavams, it has been suggested that there is sharing of intermediates between the pathways (Figure 1). Therefore, many gene products from the CA, clavam, and paralog BGCs contribute to the early part of the biosynthetic pathway involved in both CA and the 5S clavam production (Figure 1; Jensen, 2012; Hamed et al., 2013; Álvarez-Álvarez et al., 2018). Previous genetic mapping studies have shown that the BGCs for CA and Ceph-C are clustered together on the chromosomes of all CA producers to form “β-lactam superclusters” (Ward and Hodgson, 1993), but details about their sequences from S. jumonjinensis and S. katsurahamanus were lacking. It has been hypothesized that CA biosynthesis evolved in an ancestral 5S clavam producer, after it acquired the ability to produce Ceph-C by horizontal gene transfer (Challis and Hopwood, 2003). Such an arrangement leads to the coordinated biosynthesis of Ceph-C and CA, or the production of a β-lactam antibiotic and a synergistically acting β-lactamase inhibitor, respectively. The complete biosynthetic pathway leading to Ceph-C has been elucidated (Liras, 1999), but some late steps required for CA production remain unknown (Jensen, 2012; Hamed et al., 2013). Additionally, not all genes from the proposed S. clavuligerus CA BGC are required for CA production (Supplementary Table S1), and the exact function of many gene products remains to be deciphered (Jensen et al., 2004a; Valegård et al., 2013; Álvarez-Álvarez et al., 2018; Srivastava et al., 2019). Recently available genome sequences have revealed that CA-like BGCs (without any associated Ceph-C BGCs) are also present in other organisms such as Streptomyces pratensis ATCC 33331 (formerly called Streptomyces flavogriseus) and Saccharomonospora viridis DSM 43017, but neither have been shown to produce CA to date (Jensen, 2012; Álvarez-Álvarez et al., 2013). Therefore, it is still not clear as to what defines the boundaries of a functional (or minimal) CA BGC, a question that we also address in the current study.
Materials and Methods
Bacterial Strains, Plasmids, Media/Culture Conditions, and Molecular Methods
Bacterial strains and plasmids used in the current study are described in Table 1. All media/reagents were purchased from Fisher Scientific or VWR International (Canada). For routine analysis, Streptomyces cultures were maintained on International Streptomyces Project (ISP) medium 4 agar or were grown in Trypticase Soy Broth supplemented with 1% (w/v) soluble starch (TSB-S). Cultures for metabolite analysis were grown using glycerol, sucrose, proline, and glutamic acid (GSPG); starch asparagine (SA); soy; or TSB-S media (Romero et al., 1997; Tahlan et al., 2004b). All Streptomyces cultures were incubated at 28°C and liquid cultures were agitated at 250 rpm. Plasmid-bearing Streptomyces cultures were supplemented with appropriate antibiotics when required (Tahlan et al., 2004b), whereas Escherichia coli was grown and maintained as described previously (Sambrook, 2001). Standard methods were used for isolating and manipulating DNA from E. coli (Sambrook, 2001) and Streptomyces (Kieser et al., 2000; Tahlan et al., 2004b). Total RNA was isolated from S. clavuligerus grown on SA medium as described previously (Srivastava et al., 2019), and RT was performed using the Maxima H Minus First Strand cDNA Synthesis Kit (Thermo Scientific, United States). PCRs were carried out using the Phusion or Taq DNA polymerase kits (ThermoFisher, United States). When required, PCR products were cloned into the pGEM-T Easy vector (Promega, United States) according to the manufacturer’s instruction and were sequenced at the Centre for Applied Genomics, University of Toronto (Canada). All DNA oligonucleotide primers used in the study (Supplementary Table S2) were obtained from Integrated DNA Technologies (United States).
TABLE 1.
Bacterial strains and plasmids used in this study.
| Strain/plasmid | Descriptiona | Source/Reference |
| Bacterial strain | ||
| E. coli ESS | Indicator strain for Ceph-C bioassays | Wang et al., 2004 |
| E. coli ET12567/pUZ8002 | Non-methylating conjugation host carrying helper plasmid pUZ8002 (CmlR and KanR) | Kieser et al., 2000 |
| E. coli DH5α | General laboratory cloning host | Promega |
| K. pneumoniae ATCC 15380 | Indicator strain for CA bioassays (PenR) | ATCC |
| S. clavuligerus ATCC27064 | Wild-type CA producer | ATCC |
| S. clavuligerus ΔnocE | nocE null mutant | This study |
| S. clavuligerus pIJ8668-ermEp∗-nocE | Strain constitutively expressing nocE | This study |
| S. jumonjinensis NRRL 5741 | Wild-type CA producer | Jensen and Paradkar, 1999 |
| S. katsurahamanus T272 | Wild type CA producer | Jensen and Paradkar, 1999 |
| Plasmids | ||
| pGEMT-Easy | Plasmid for cloning PCR products | Promega |
| pIJ8668-ermEp∗ | Conjugative Streptomyces suicide vector containing ermEp∗ for chromosomal promoter insertion (AprR) | (Tahlan et al., 2017) |
| pIJ8668-ermEp∗-nocE | pIJ8668-ermEp∗ containing a portion of the 5′ end of nocE from S. clavuligerus (AprR) | This study |
| pIJ12738 | Conjugative Streptomyces suicide vector containing an I-SceI site for gene targeting (AprR) | Fernández-Martínez and Bibb, 2014 |
| pIJ12738-nocE-UP-DN | pIJ12738 containing regions upstream and downstream of nocE from S. clavuligerus (AprR) | This study |
| pIJ12742 | Plasmid expressing the Meganuclease I-SceI in Streptomyces for gene disruption (AprR and TsrR) | Fernández-Martínez and Bibb, 2014 |
aAprR, apramycin resistance; CmlR, chloramphenicol resistance; KanR, kanamycin resistance; PenR, penicillin G resistance; and TsrR, thiostrepton resistance.
Genome Sequencing, Gene Cluster Identification, and Bioinformatics Analyses
The S. jumonjinensis and S. katsurahamanus genomes were sequenced using Illumina MiSEQ in paired-end format with read lengths of 300 bp. A chromosomal DNA library was prepared for each organism using the PCR-based method adjusted for high GC DNA according to the manufacturer’s instructions (Illumina, United States). Raw reads were filtered with trimmomatic (Bolger et al., 2014) with a cutoff of 26 bp and a minimum length of 150 bp. The remaining reads were assembled using Velvet (Zerbino and Birney, 2008). k-mers from 30 to 170 were tested for selecting optimal contig length and the assembled genomes (31–46 × coverage, Supplementary Table S3) were submitted to NCBI (accession numbers: S. jumonjinensis NRRL 5741, VCLA00000000 and S. katsurahamanus T-272, VDEQ00000000). Genome completeness was calculated (Supplementary Table S3) using BUSCO (Simao et al., 2015) and QUAST (Gurevich et al., 2013). Annotations were carried out using RAST (Overbeek et al., 2014) and also manually in Artemis (Rutherford et al., 2000). Specialized metabolite (SM) biosynthetic gene clusters (BGCs) were identified using antiSMASH 4.0 (Blin et al., 2017) and polyketide synthases/nonribosomal peptide synthetase genes were predicted using PRISM 3 (Skinnider et al., 2017). The DNA sequences of S. jumonjinensis and S. katsurahamanus BGCs were manually examined for possible frame shifts and other ambiguities. In some cases, PCR amplification was performed using custom primers (Supplementary Table S2) followed by Sanger sequencing of products to verify results. The genome sequences of S. clavuligerus ATCC 27064 (NZ_CM000913.1, NZ_CM000914.1), S. pratensis ATCC 33331 (NC_016114), S. viridis DSM 43017 (CP001683.1), Streptomyces sp. M41(2017) (NZ_MWFK00000000), Streptomyces sp. PAMC26508 (NC_021055), Streptomyces sp. NRRL S-325 (NZ_JOIW00000000), Streptomyces sp. NRRL B-24051 (NZ_JOAE00000000), Streptomyces flavovirens NRRL B-2182 (NZ_JOAB00000000), Streptomyces fulvoviridis NRRL ISP-5210 (NZ_JNXH00000000), and Streptomyces olivaceus NRRL B-3009 (NZ_JOFH00000000) were included for comparison as the latter harbor CA-like BGCs containing homologs of all genes currently known to be involved in CA production in S. clavuligerus (Jensen, 2012). In addition, the sequences of the Ceph-C BGCs from Streptomyces cattleya 8057 (NC_017586.1) and Nocardia lactamdurans (also known as Amycolatopsis lactamdurans) (Z13971.1-Z13974.1, Z21681.1-Z21686.1 and X57310.1) were also included in the analysis. Geneious 8.1.9 (Biomatters Ltd., New Zealand) was used for sequence comparisons and constructing phylogenetic trees. Protein homologs were identified using NCBI BLAST and secretory signals were predicted using the SignalP-5.0 Server (Almagro Armenteros et al., 2019).
Preparation of the S. clavuligerus ΔnocE and ermEp∗-nocE Strains
The S. clavuligerus nocE gene mutant was prepared using the meganuclease I-SceI marker-less gene deletion system (Fernández-Martínez and Bibb, 2014). DNA fragments (1–1.2 kb each) containing regions immediately upstream and downstream of nocE from the S. clavuligerus chromosome were amplified using PCR along with engineered primers (Supplementary Table S2) and were separately cloned into the pGEM-T Easy vector (Table 1). The upstream fragment was released from pGEM-T Easy by digestion with HindIII and EcoRI and was introduced into the same sites of pIJ12738 to give pIJ12738/nocE-UP. The downstream fragment was then introduced into the EcoRI and XbaI sites of pIJ12738-nocE-UP to give pIJ12738/nocE-UP-DN, which functioned as the nocE disruption construct (Table 1). pIJ12738-nocE-UP-DN was conjugated into S. clavuligerus to obtain the apramycin-resistant single crossover strain, which was confirmed using genomic DNA PCR (Supplementary Table S2). The plasmid pIJ12742 expressing the I-SceI meganuclease (Table 1) was then conjugated into S. clavuligerus pIJ12738-nocE-UP-DN to obtain apramycin and thiostrepton resistant exconjugants, which were made to undergo sporulation at 28°C without any selection to facilitate double homologous recombination and loss of pIJ12738 from the chromosome. Spore stocks were prepared and re-streaked onto ISP-4 plates without selection and incubated for 5 days at 37°C to promote the loss of temperature-sensitive pIJ12742. This led to the isolation of the apramycin and thiostrepton-sensitive S. clavuligerus ΔnocE mutant, which was verified using genomic DNA PCR (Supplementary Table S2).
To prepare an S. clavuligerus strain constitutively expressing nocE (Table 1), the ermEp∗ promoter (Bibb et al., 1985) was inserted upstream of the gene in the S. clavuligerus chromosome. A 1.1-kb DNA fragment from the 5′ end of the gene was amplified by PCR (Supplementary Table S2) and was cloned into pGEM-T Easy. The insert was re-isolated as an NdeI and EcoRI fragment and was ligated with similarly digested pIJ8668-ermEp∗ to give pIJ8668-ermEp∗-nocE (Table 1), which was introduced into wt S. clavuligerus by conjugation. This resulted in the S. clavuligerus ermEp∗-nocE strain, which was confirmed using genomic DNA PCR (Supplementary Table S2) and was used to examine the effect of constitutively expressing nocE in S. clavuligerus.
Bioassays and Bacterial Growth Measurement
The production of CA and Ceph-C in culture supernatants was detected (and quantified in the case of Ceph-C) using bioassays employing indicator organisms (Table 1), as described previously (Paradkar and Jensen, 1995; Wang et al., 2004). Growth in liquid cultures was determined using a modified diphenylamine colorimetric method to measure DNA content (Zhao et al., 2013) and statistical analysis (ANOVA repeated measure) was performed using R 3.4.3. To assess for growth characteristics on solid media, 10-fold dilutions of a spore stock (4 × 104 spores/μl) were prepared, and 5 μl of which were spotted onto two different agar media (SA and TSB-S with 1.5% agar). The plates were then incubated at 28°C and visually scored for growth over a 7-day period.
Liquid Chromatography–Mass Spectrometry (LC-MS and LC-MS/MS) Analysis
The production of clavam metabolites in 96-h broth cultures was analyzed by targeted LC-MS after imidazole derivatization using an XTerra column (2.1 × 150 mm, 3.5 μm, 125 Å; Waters Scientific, United States) as described previously (Srivastava et al., 2019). Untargeted metabolomics was conducted using bacteria grown on solid media. One hundred microliters of a standardized spore stock (4 × 104 spores/μl) of each species was used to inoculate agar plates in duplicate, and each plate was extracted using 15 ml of methanol or ethyl acetate. Two milliliters of each extract was dried, resuspended in 130 μl of 70% methanol containing 0.2 μM of amitriptyline (internal standard), and transferred to a 96-well plate, which was centrifuged at 2000 rpm for 15 min at 4°C. One hundred microliters of each sample was then transferred to a new 96-well plate for LC-MS/MS analysis. Samples were analyzed using a Vanquish UHPLC System coupled Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Scientific, United States). Chromatographic separation was performed in mixed mode (allowing weak anion/cation exchange) on a Scherzo SM-C18 column (2 × 250 mm, 3 μm, 130 Å; Imtakt, United States) maintained at 40°C. Ten microliters of each sample was injected for analysis and the mobile phase consisted of (A) 0.1% formic acid in water and (B) 0.1% formic acid in acetonitrile. Chromatography was performed at a flow rate of 0.5 ml/min using the following program: 0–5 min, 98% A; 5–8 min, gradient of 98–50% A (or 50% B); 8–13 min, gradient 50–100% B; 13–14.00 min, 100% B; 14–14.10 min, 100–2% B; 14.10–18 min, 2% B.
Mass spectrometry was performed using a heated electrospray ionization source (heater temperature, 370°C and capillary temperature, 350°C) in either positive or negative ionization mode (± 3000.0 V; S-lens RF, 55; sheath gas flow rate, 55; and auxiliary gas flow rate, 20). MS1 and MS2 scans (at 200 m/z) were acquired from 0.48 to 16.0 min at a resolution of 35,000 and 17,500, respectively, for the 100–1500 m/z range. The automatic gain control (AGC) target value and maximum injection time were set at 5 × 105 and 150 ms. Up to four MS2 scans in data-dependent mode were acquired for most abundant ions per duty cycle, with a starting value of 70 m/z, and exclusion parameter of 10 s. Higher-energy collision-induced dissociation was performed with a normalized collision energy of 20, 35, and 50 eV. The apex trigger mode was used (2–7 s) and the isotopes were excluded. Inclusion lists of ions for molecules observed in Streptomyces extracts were generated from the Dictionary of Natural Products1 and the StreptomeDB (Lucas et al., 2013), and were used for prioritizing the acquisition of their MS2 when observed. The raw LC-MS/MS data files were converted to .mzXML format using ProteoWizard (Adusumilli and Mallick, 2017). All metabolomics MS data have been deposited on the MassIVE public repository2 under the accession number MSV000083835.
MS Data Annotation and Analysis
Molecular networks were generated using positive and negative ionization mode data in GNPS (Wang et al., 2016). The resulting networks were visualized in Cytoscape (Shannon et al., 2003), allowing nodes associated with uninoculated media controls to be removed. Annotations were first obtained by matching spectra in public libraries (Wang et al., 2016), including NIST173. Library annotations were manually validated using mirror plots (maximum ion mass accuracy = 5 ppm) corresponding to level 2 annotation based on the Minimum Standard Initiative (Spicer et al., 2017). The data were deposited to the GNPS library (CCMSLIB00005435954-CCMSLIB00000531493), which enabled the annotation of putative tunicamycin derivatives (CCMSLIB00005435941-42) and lyngbyatoxin (CCMSLIB00005435954-55) using molecular networks. In some cases, Sirius 4.0.1 was used to confirm the molecular formulas of certain predicted metabolites (Böcker et al., 2009).
To generate a heat map using the S. clavuligerus wt, ΔnocE, and ermEp∗-nocE strains, feature-based detection and alignment of positive mode ionization data were performed (parameters: MS1 noise level of 25000, MS2 noise level of 1000) using the MZmine 2 toolbox (v2.39) (Pluskal et al., 2010). Chromatograms were built using the ADAP module (parameters: min group size in # of scans = 4, group intensity threshold = 700,000, min highest intensity = 100,000, max m/z tolerance = 10 ppm), which were then deconvoluted (parameters: S/N threshold = 10.0, min feature height = 7000000, coefficient/area threshold = 60.0, peak duration range = 0.01–0.5 min, RT wavelet range = 0.01–0.1 s). Fragmentation spectra were paired with deconvoluted peaks using 0.02 Da and 0.2 min windows, and LC-MS features were annotated using the Peak-Grouping module (parameters: deisotope = true, remove features without isotope pattern = false, minimal intensity for interval selection = 0.1, minimal intensity overlap = 0.7, minimal correlation = 0.7). Features were aligned in the JoinAligner module (parameters: ppm tolerance = 7, weight for m/z = 75.0, retention time tolerance = 0.5 min, weight for RT = 25.0; require same charge state = false, require same ID = false, compare isotope pattern = false). The aligned peaklist was filtered with the row filter module to keep only features with at least two isotopic ions, two occurrences, and at least one MS2 spectrum before gap filling (parameters: intensity = 5%, ppm window = 5, retention time tolerance = 0.15). The aligned peaklist containing 3149 features was exported as a .CSV file, and the spectral data as .MGF files using the GNPSExport module for further processing. The signal intensities of the features (.CSV) were normalized to that of an internal standard (m/z 278.189; retention time, 9.2 min) and only 1684 features with an intensity 3-fold higher than in experimental controls (uncultivated media) were retained. MetaboAnalyst4.0 (Chong et al., 2018) was used to perform the hierarchal clustering, which was visualized as a heat map.
Results and Discussion
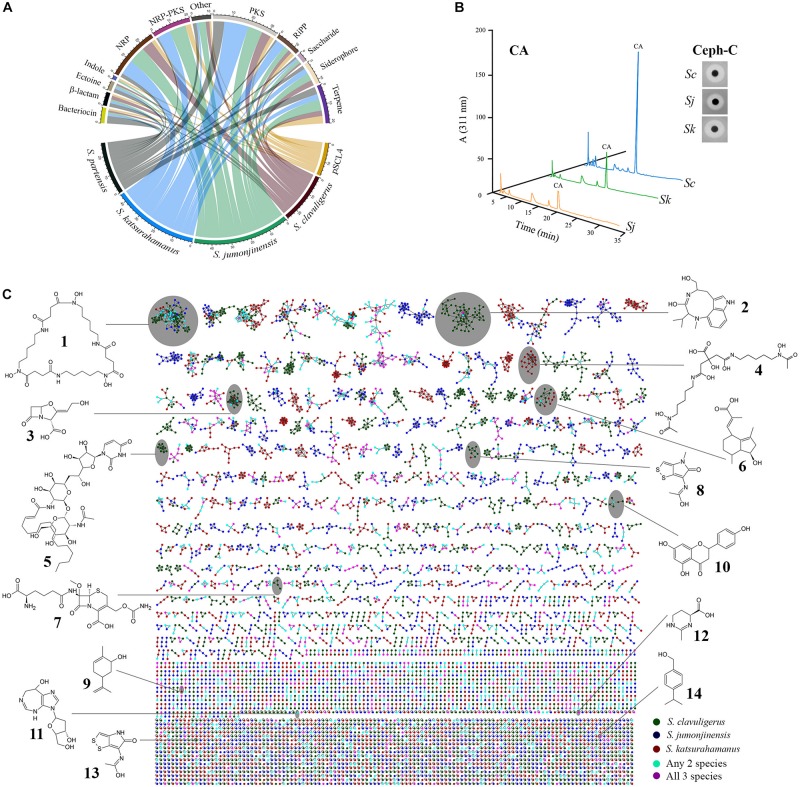
Three Streptomyces species are known to produce CA, but details about the involved BGCs are only available for the genome sequenced industrial producer, S. clavuligerus (Medema et al., 2010; Song et al., 2010b; Cao et al., 2016). Therefore, we sequenced the genomes of the other two CA producers, S. jumonjinensis and S. katsurahamanus (Table 1), for comparative studies. The published genome sequence of S. pratensis ATCC 33331 was also included during some of the analyses (Figure 2A), as it contains a CA-like BGC (Figures 3A,C), and has been shown not to produce the metabolite under tested conditions (Álvarez-Álvarez et al., 2013). Examination of the S. jumonjinensis and S. katsurahamanus genomes revealed that they each contain 49 and 44 known or predicted SM BGCs (Table 2 and Supplementary Table S4), respectively, which is much higher than the average number found in many Streptomyces species. Additionally, S. clavuligerus contains 43 SM BGCs, although re-sequencing of its genome suggests that it may contain many more (Hwang et al., 2019). This prompted us to further investigate the specialized metabolic capabilities of the three CA producers to determine similarities or differences between these microorganisms.
FIGURE 2.
Biosynthetic gene cluster (BGC) content and metabolomics analysis of clavulanic acid (CA)-producing Streptomyces species (S. clavuligerus, S. jumonjinensis, and S. katsurahamanus). (A) Circular chord diagram representing all predicted BGC classes present in the respective Streptomyces species. S. pratensis was included for comparison as the bacterium contains a CA-like BGC, but does not produce the metabolite. The sequence of the giant linear plasmid pSCL4 from S. clavuligerus was also included separately due to the presence of multiple BGCs on it. The lower arc represents genomes/plasmid, while the upper arc represents different classes of BGCs and the color-coded ribbons connecting them indicate the presence of a BGC in the specific species. (B) Detection of CA and cephamycin C (Ceph-C) in 96-h SA culture supernatants of S. clavuligerus (Sc), S. jumonjinensis (Sj), and S. katsurahamanus (Sk) using LC-MS (after imidazole derivatization) and bioassays (inset), respectively. The peak corresponding to CA in HPLC chromatograms is noted and the zones of inhibition in the inset panel demonstrate relative amounts of Ceph-C production. (C) Metabolic network constructed using S. clavuligerus, S. jumonjinensis, and S. katsurahamanus culture extracts (culture conditions and details are described in the section “Materials and Methods”). The network is color-coded according to source organism (bottom right legend), where each node depicts a mass spectrum and edges represent the relationship between different nodes. Structures of natural products detected in the extracts at high confidence in the three species are shown and the clade in the network containing the node corresponding to the respective metabolite is also indicated. 1, desferrioxamine E; 2, (−)-indolactam V; 3, clavulanic acid; 4, arthrobactin; 5, tunicamycin C2; 6, hydroxyvalerenic acid; 7, cephamycin C; 8, thiolutin; 9, (−)-carveol; 10, naringenin; 11, pentostatin, 12, ectoine; 13, holomycin; and 14, cuminyl alcohol.
FIGURE 3.
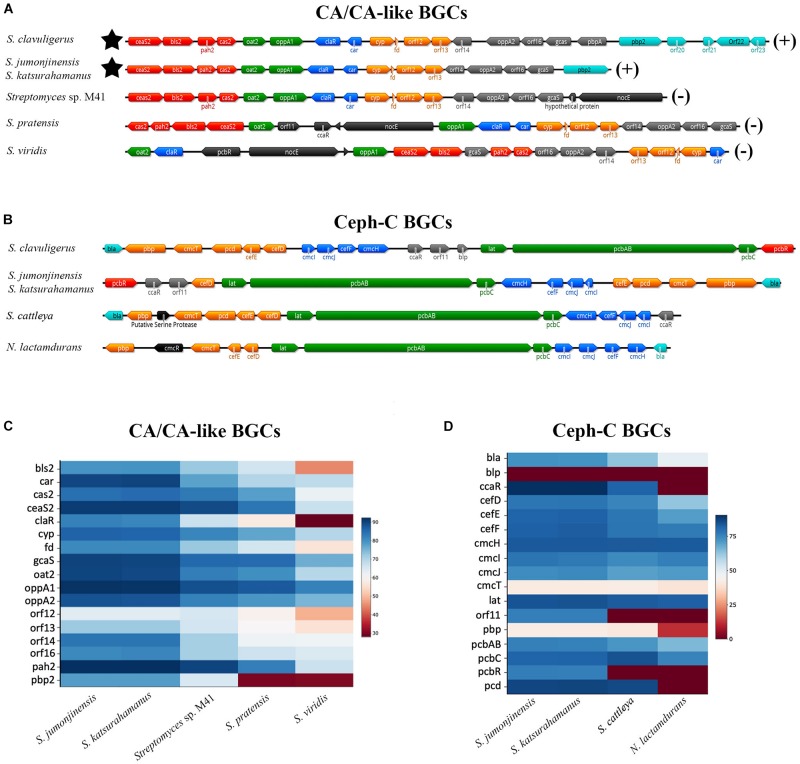
Organization of CA/CA-like (A) and Ceph-C (B) BGCs in select CA-producing/non-producing Streptomyces species. The CA non-producers Saccharomonospora viridis (A,C) and Nocardia (or Amycolatopsis) lactamdurans (B,D) were also included for comparison as both are phylogenetically distinct from the Streptomyces. (A,B) The architecture of respective BGCs from the described organisms showing their gene content and relative organization. Genes are color-coded based on known or predicted transcriptional units. (A) The star symbol represents the location of the Ceph-C BGC (if present) and the CA production status (±) of each organism is indicated on the right. (B) All species included are Ceph-C and CA producers except for S. cattleya and N. lactamdurans, which only produce the former. (C,D) Relative identities of protein products from the CA/CA-like (C) and Ceph-C (D) BGCs of described organisms as compared to corresponding homologs from S. clavuligerus. The legend on the right shows colors indicating percent identities between respective gene products.
TABLE 2.
Genome features relevant to specialized metabolism in S. jumonjinensis and S. katsurahamanus as compared to S. clavuligerus (CA producer) and S. pratensis (CA non-producer).
| Feature | S. jumonjinensis NRRL 5741 | S. katsurahamanus T272 | S. clavuligerus ATCC27064 | S. pratensis ATCC 33331 |
| Genome size (Mbp) | 8.47a | 7.25a | 8.56 | 7.34 |
| Coding sequences | 7423 | 6123 | 7281 | 6537 |
| SM BGCsb (PKS/NRPS)c | 49 (8/18) | 44 (9/9) | 43 (10/9) | 27 (5/9) |
a Estimated sizes based on sequence analysis from the current study. b Specialized metabolite (SM) biosynthetic gene clusters (BGCs) were predicted using antiSMASH 4.0. c Polyketide synthase (PKS) and nonribosomal peptide synthetase (NRPS) genes were predicted using PRISM 3.
SM-BGCs and Metabolism in S. clavuligerus, S. jumonjinensis, and S. katsurahamanus
Detailed analysis of the S. jumonjinensis and S. katsurahamanus genome sequences using antiSMASH 4.0 (Blin et al., 2017) and manual curation showed that both organisms contain numerous BGCs for diverse SMs (Figure 2A and Supplementary Table S4). Therefore, S. clavuligerus, S. jumonjinensis, and S. katsurahamanus were grown on SA, GSPG, and TSB-S media for assessing CA/Ceph-C production (Figure 2B and Supplementary Figure S1) and for preparing methanol/ethyl acetate extracts for liquid chromatography–tandem mass spectrometry (LC-MS/MS)-based metabolomics. The MS/MS data obtained from both positive and negative ionization mode were used to build a molecular network (Figure 2C), and metabolites were annotated by matching spectra against public libraries corresponding to level 2 annotation based on the Metabolomics Standard Initiative (Wang et al., 2016). During the analysis, ions corresponding to CA ([M-H]–, m/z 198.039), Ceph-C ([M-H]–, m/z 445.104) and numerous other SMs were also detected in extracts from one or more Streptomyces species (Figure 2C, Supplementary Tables S5, S6), some of which are discussed below.
The desferrioxamines (Figure 2C) comprise a group of nonpeptide hydroxamate siderophores produced by many bacteria (Barona-Gómez et al., 2004), including S. clavuligerus (Álvarez-Álvarez et al., 2017). In the current study, ions corresponding to desferrioxamine E (Nocardamine, [M + H]+, m/z 601.356) and desferrioxamine B (Desferal, [M + H]+, m/z 561.361) were detected in extracts from S. clavuligerus/S. jumonjinensis and S. clavuligerus/S. katsurahamanus, respectively (Supplementary Table S5). Desferrioxamine E exhibits antitumor activity (Kalinovskaya et al., 2011), while desferrioxamine B is used in therapy for secondary iron overload disease (Olivieri and Brittenham, 1997). We also identified BGCs in S. clavuligerus, S. jumonjinensis, and S. katsurahamanus (Supplementary Table S4) that have high degrees of similarity (80–100%) with BGCs from known desferrioxamine producers such as Streptomyces griseus (Yamanaka et al., 2005; Ohnishi et al., 2008) and Streptomyces coelicolor A3(2) (Bentley et al., 2002; Barona-Gómez et al., 2004). The siderophore arthrobactin (Figure 2C) was also detected in S. katsurahamanus extracts ([M + H]+, m/z 477.256) (Supplementary Table S5), but since the genes responsible for arthrobactin production are not known (Burrell et al., 2012), we were unable to identify an associated BGC in this organism. However, our analysis showed that S. clavuligerus, S. jumonjinensis, and S. katsurahamanus each contain additional siderophore-like BGCs of unknown function (Supplementary Table S4), which could potentially be involved in the production of such metabolites. Ectoine is another commonly produced metabolite that helps bacteria survive extreme osmotic stress (Sadeghi et al., 2014), and it was detected ([M + H]+, m/z 143.082) in extracts from all three CA-producing species (Figure 2C and Supplementary Table S5). In addition, S. clavuligerus, S. jumonjinensis, and S. katsurahamanus contain BGCs that are similar to the known ectoine BGC from Streptomyces anulatus (previously called Streptomyces chrysomallus) (Prabhu et al., 2004). Since the desferrioxamines and ectoine are produced by many Actinomycetes and are involved in general cellular growth/survival processes (Challis, 2005; Czech et al., 2018), finding them in culture extracts from the three CA producers in the current study was not surprising.
Streptomyces clavuligerus is a known producer of the dithiolopyrrolone antibiotic holomycin (Kenig and Reading, 1979) and the associated BGC has been identified in this organism (Li and Walsh, 2010). In the current study, holomycin ([M + H]+, m/z 214.994) and thiolutin (another dithiolopyrrolone, [M + H]+, m/z 229.010) were detected in extracts from S. clavuligerus, but not in those from S. jumonjinensis or S. katsurahamanus (Figure 2C and Supplementary Table S5). Recently, a dithiolopyrrolone with the same molecular weight as thiolutin (predicted to be N-propionylholothin) was also detected in extracts from S. clavuligerus strains lacking the giant linear plasmid pSCL4 (Álvarez-Álvarez et al., 2017). Since holomycin and thiolutin (Figure 2C), and the respective BGCs involved in their biosynthesis (from S. clavuligerus and Saccharothrix algeriensis NRRL B-24137, respectively), are very similar (Supplementary Table S4), it is possible that a single pathway in S. clavuligerus produces both metabolites. It has also been reported that there is some sort of cross regulation between CA and holomycin production in S. clavuligerus (de la Fuente et al., 2002; Álvarez-Álvarez et al., 2017). Our results showed that S. jumonjinensis and S. katsurahamanus lack dithiolopyrrolone BGCs (Supplementary Table S4) and therefore do not have a similar link between holomycin and CA production as observed in S. clavuligerus.
We also detected certain nucleoside SMs during the current analysis (Figure 2C). For example, the purine nucleoside pentostatin, which is also used as an anticancer agent (Dillman, 2004), was identified ([M + 2H]2+, m/z 135.066) in S. clavuligerus extracts (Figure 2C, Supplementary Table S5). A putative pentostatin-like BGC was recently shown to be present in S. clavuligerus (Wu et al., 2017), but production of the metabolite has not been reported in this organism previously. Therefore, our results suggest that the S. clavuligerus pentostatin BGC can be activated under laboratory conditions. The tunicamycins also comprise a mixture of related nucleoside antibiotics, some of which (A, B, C, and I) were detected in extracts from S. clavuligerus (Figure 2C and Supplementary Table S5), but not in those from S. jumonjinensis or S. katsurahamanus. S. clavuligerus is a known producer of tunicamycin and the BGC involved in its production has been identified (Kenig and Reading, 1979; Chen et al., 2010). In addition, certain derivatives of tunicamycin I with different acyl chains were detected in S. clavuligerus extracts recently (Martínez-Burgo et al., 2019), which were also present in our samples (Supplementary Table S5). Our results demonstrated that S. jumonjinensis and S. katsurahamanus do not possess tunicamycin BGCs (Supplementary Table S4), further distinguishing S. clavuligerus from the other CA producers due to its ability to produce such nucleoside SMs.
Metabolomics analysis also revealed the presence of certain plant-associated SMs in the Streptomyces extracts. It was recently shown that S. clavuligerus produces the citrus flavonoid naringenin and the genes involved in the production of this metabolite were also identified (Álvarez-Álvarez et al., 2015). Naringenin exhibits antibacterial, antifungal, and anticancer activities (Rauha et al., 2000; Kanno et al., 2005), and its production by a bacterium was unexpected since it was previously isolated from plants only (Álvarez-Álvarez et al., 2015). We detected naringenin (Figure 2C, [M-H]–, m/z 271.062) in extracts from S. clavuligerus and S. jumonjinensis, but not from S. katsurahamanus (Supplementary Table S5). In addition, the genes involved in naringenin production were also found in both S. jumonjinensis and S. katsurahamanus (Supplementary Table S4), suggesting that the metabolite might be produced at undetectable levels in S. katsurahamanus or that the genes are not expressed in this species under the conditions tested. Also detected in all three Streptomyces extracts were the plant-associated monoterpenes, carveol ([M-H2O + H]+, m/z 135.117), and cuminyl alcohol ([M-H2O + H]+, m/z 133.101), whereas hydroxyvalerenic acid (another plant terpene, [2M-H]–, m/z 499.307) was found in S. clavuligerus extracts only (Figure 2C and Supplementary Table S5). The pathways involved in the production of the latter three metabolites are not fully known (Wong et al., 2018), however, S. clavuligerus, S. jumonjinensis, and S. katsurahamanus possess many terpene-like BGCs of unknown function, which could potentially be involved in their biosynthesis (Supplementary Table S4). Therefore, our results suggest that certain Streptomyces also harbor the capacity to produce carveol, cuminyl alcohol, and hydroxyvalerenic acid along with naringenin, a finding that can be potentially exploited for further development.
The indole alkaloid, (−)-indolactam V is a protein kinase C activator (Heikkila and Akerman, 1989) and functions as an intermediate during the biosynthesis of other SMs in certain Actinomycetes (Abe, 2018). We detected (−)-indolactam V (Figure 2C, [M-CO + H]+, m/z 274.191) and some of its alkylated derivatives in extracts from S. clavuligerus, but not in those from S. jumonjinensis or S. katsurahamanus (Supplementary Table S5). The genes normally associated with (−)-indolactam V biosynthesis could not be identified in the current study, warranting further investigation into its production in S. clavuligerus. Other metabolites were also detected during the analysis (Supplementary Table S6), but we were unable to find details about their biosynthesis in bacteria or predict associated BGCs, and therefore we did not include them in the discussion. In addition, S. jumonjinensis and S. katsurahamanus contain several BGCs related to known pathways for which products could not be detected (Supplementary Table S4). For example, there is an NRPS-containing BGC in S. jumonjinensis that is 100% similar to the BGC in Streptomyces sp. DSM 11171, which produces the antiviral metabolite feglymycin (Supplementary Table S4; Gonsior et al., 2015). We also identified indole-associated BGCs in S. clavuligerus and S. jumonjinensis (Supplementary Table S4), which are similar to the one from Streptomyces sp. TP-A0274 responsible for producing the anticancer agent staurosporine (Onaka et al., 2002). Similarly, BGCs for polycyclic tetramate macrolactams (PTMs, NRP/PKs) are present in both S. jumonjinensis and S. katsurahamanus, which are 100% similar to a SGR-PTM BGC from the known producer S. griseus (Supplementary Table S4; Luo et al., 2013). PTMs possess antifungal and antioxidant properties, and cryptic PTM-like BGCs are commonly found in Streptomyces genomes (Zhang et al., 2016). Moreover, BGCs for many other classes of SMs including enediynes (Rudolf et al., 2016) and the ribosomally synthesized and post-translationally modified peptides (RiPPs) (Hetrick and van der Donk, 2017) were also identified in S. jumonjinensis and S. katsurahamanus (Supplementary Table S4), but further work is required to detect their production in these organisms. In the current study, >14,000 molecular nodes were obtained using MS-based metabolomics and GNPS analysis (Figure 2C), but only 10% could be annotated by matching spectra with available libraries. Therefore, many of the unannotated nodes could represent products of so-called “cryptic” BGCs, a situation that should change over time as databases are populated with more spectra from authentic samples.
Comparative Sequence Analysis of CA-BGCs From Streptomyces Species
In addition to analyzing the overall SM production capabilities of CA producers, we were also interested in specifically examining the BGCs involved in β-lactam biosynthesis from S. jumonjinensis and S. katsurahamanus for comparison with S. clavuligerus (Figure 3). The genome sequences of S. jumonjinensis and S. katsurahamanus revealed that they both contain identical CA and Ceph-C BGCs (Figure 3), but lack the clavam and paralog gene clusters (Supplementary Table S4). This would explain why they do not produce the 5S clavams as compared to S. clavuligerus (Jensen, 2012). The results further confirm that intact 5S clavam and paralog BGCs are not essential for CA production (Figure 1), since both S. jumonjinensis and S. katsurahamanus can produce the metabolite (Figure 2B and Supplementary Figure S1). The paralog gene cluster from S. clavuligerus contains second copies of certain genes (ceaS1, bls1, and pah1) from the CA BGC (Jensen et al., 2004b; Tahlan et al., 2004b), which encode enzymes involved in the early shared stages of CA and 5S clavam biosynthesis (Figure 1). It has also been shown that the remaining un-duplicated genes from the paralog gene cluster and almost all genes from the clavam gene cluster (except one; cas1) are exclusively involved in 5S clavam production (Mosher et al., 1999; Tahlan et al., 2007; Zelyas et al., 2008). Therefore, our results provide additional support for the hypothesis that the clavam and paralog gene clusters are associated with 5S clavam biosynthesis, and that some gene products from the two clusters augment CA production in S. clavuligerus by contributing to a common pool of precursors (Figure 1; Jensen, 2012; Hamed et al., 2013). Although, it should be noted that in S. clavuligerus, there is some cross regulation between the chromosomal CA and plasmid-borne paralog gene clusters (Kwong et al., 2013; Álvarez-Álvarez et al., 2017), which is again not expected to occur in the other two CA producers since they only contain the CA BGC. This also highlights the complexity of the regulatory pathways controlling CA and 5S clavam production in S. clavuligerus (Liras et al., 2008). For this reason, we focused our analysis and discussion on the comparison of biosynthetic genes (and BGCs), instead of regulation. In the current study, CA production levels in S. jumonjinensis and S. katsurahamanus could never match those observed in wt S. clavuligerus, whereas all three species produced Ceph-C at comparable levels (Figure 2B). It has been previously suggested that higher CA yields in S. clavuligerus might be explained in part by increased precursor supply for biosynthesis due to the presence of the paralog and clavam gene clusters in this species (Figure 1). In addition, enhanced levels of biosynthetic gene expression could be another reason why S. clavuligerus is currently the preferred industrial producer and was first identified in screens for β-lactamase inhibitors, as higher CA yields would make it easier to detect during assays (Jensen, 2012).
Closer examination of the CA BGCs from S. jumonjinensis and S. katsurahamanus showed that they each contain most of the genes from the corresponding S. clavuligerus BGC in the same order, except that orf18 (pbpA), orf20, orf21, orf22, and orf23 are absent (Figure 3A). pbpA is predicted to encode a high-molecular-weight penicillin-binding protein (PBP), but its role in CA production remains unknown (Jensen et al., 2004a). Previous studies have also shown that disruption of orf19 (pbp2) (Jensen et al., 2004a), orf20 (cytochrome P-450) (Song et al., 2009), orf21 (putative sigma factor), orf22 (sensor kinase), or orf23 (response regulator) (Fu et al., 2019a) in S. clavuligerus does not abolish CA or Ceph-C production (Song et al., 2009; Supplementary Table S1). Since the respective genes are not present in S. jumonjinensis and S. katsurahamanus (Figure 3A), it is apparent that they are not part of the core BGC required for biosynthesis, but instead have accessory roles in S. clavuligerus. In a previous study, it was also shown that the expression of orf18–21 was not significantly affected in a S. clavuligerus mutant defective in ClaR, the cluster-situated regulator responsible for controlling CA biosynthesis (Martínez-Burgo et al., 2015). Therefore, we propose that the core CA BGC comprises ceaS2 (encoding carboxyethylarginine synthase), gcas (encoding N-glycyl-clavaminic acid synthetase), and the intervening genes (Figure 3A, and Supplementary Table S1).
The CA and Ceph-C BGCs in S. jumonjinensis and S. katsurahamanus also form “β-lactam superclusters” as observed in S. clavuligerus, which agrees with previous restriction mapping studies (Ward and Hodgson, 1993). The linkage of the Ceph-C and CA BGCs in S. clavuligerus, S. jumonjinensis, and S. katsurahamanus, and the coordinated production of the two metabolites in S. clavuligerus (Pérez-Llarena et al., 1997), provides further evidence for the simultaneous acquisition of the two BGCs by producing species. It has been proposed that the CA BGC might have evolved by the duplication of an ancestral 5S clavam BGC and the acquisition of the ability to produce Ceph-C in the same organism (Challis and Hopwood, 2003). Such a situation led to the selection for the ability to produce a β-lactamase inhibitor, resulting in the assembly of the currently known CA BGC, and the formation of the β-lactam supercluster (Challis and Hopwood, 2003). Our results showed that the Ceph-C BGCs from S. jumonjinensis and S. katsurahamanus are identical to each other, but differ slightly from those present in S. clavuligerus and other Ceph-C-producing Actinobacteria (Figures 3B,D). The positions of genes forming individual operons (or transcriptional units) in all three CA producers is very similar (except for the location of cefD), but the relative arrangement of operons is different in S. jumonjinensis and S. katsurahamanus as compared to S. clavuligerus (Figure 3B). In addition, the Ceph-C BGCs of S. jumonjinensis, S. katsurahamanus and other previously reported Ceph-C producers (other than S. clavuligerus) (Liras et al., 1998) do not contain blp (Figure 3B), which encodes a product resembling β-lactamase inhibitory proteins (Blip), but has been shown to lack any such activity (Gretes et al., 2009). Previous studies have shown that disruption of blp does not affect Ceph-C or CA production in S. clavuligerus (Alexander and Jensen, 1998; Thai et al., 2001). Therefore, blp does not seem to a part of the core Ceph-C BGC since S. jumonjinensis, S. katsurahamanus, and other species shown in Figure 3B can still produce the metabolite in its absence. Another noticeable feature of Ceph-C BGCs from the three CA producers is the presence of pcbR, which is missing from the homologous BGCs of species that only produce Ceph-C, but not CA (Figure 3B). PcbR resembles PBPs (Paradkar et al., 1996), but it is not essential for Ceph-C biosynthesis since it is not present in the BGCs of all organisms capable of producing the metabolite (Figure 3B, more details below).
Overall, the “β-lactam superclusters” from S. clavuligerus, S. jumonjinensis, and S. katsurahamanus are very similar to each other (Figures 3C,D). In comparison, CA-like BGCs from non-producers are markedly different, and do not form “β-lactam superclusters” as they lack Ceph-C BGCs (Jensen, 2012). The non-producers (including some Streptomyces) are also phylogenetically distinct from CA-producing species (Supplementary Figure S2), and their CA-like BGCs show three distinct patterns in terms of gene content and arrangement (Figure 3A). Many organisms in the database contain CA-like BGCs identical to the one found in S. pratensis, whereas we could only find one example each of the types present in Streptomyces sp. M41 and S. viridis, respectively (Figure 3A). In addition, CA-like BGCs from S. pratensis and S. viridis contain the pcbR, orf11, and nocE genes (Álvarez-Álvarez et al., 2013), which are not present in the CA BGCs of S. clavuligerus, S. jumonjinensis, or S. katsurahamanus (Figure 3A). Interestingly, pcbR and orf11 are included in the Ceph-C BGCs of CA producers, whereas nocE is located elsewhere on the chromosome in the three Streptomyces species (Figure 3B). As mentioned earlier, pcbR encodes a PBP involved in β-lactam resistance (Paradkar et al., 1996), whereas orf11 encodes a predicted protein of unknown function. Previous reports have shown that disruption of neither pcbR nor orf11 in S. clavuligerus affected Ceph-C or CA production (Paradkar et al., 1996; Alexander and Jensen, 1998), suggesting that they are not required for the biosynthesis of the respective metabolites.
The presence of nocE homologs in CA producers and in the CA-like BGCs of all non-producers is intriguing (Figure 3A), as they are similar to a gene from the nocardicin A monobactam BGC of Nocardia uniformis (Gunsior et al., 2004). The nocE genes are predicted to encode proteins containing C-terminal SGNH/GDSL hydrolase family domains, which are normally associated with esterases or lipases (Upton and Buckley, 1995), but their function during β-lactam metabolite biosynthesis is not obvious. The disruption of nocE in N. uniformis does not affect nocardicin A production (Davidsen and Townsend, 2009), but the role of the gene in β-lactam-producing Streptomyces has not been examined to date.
Examination of the Function of nocE in S. clavuligerus
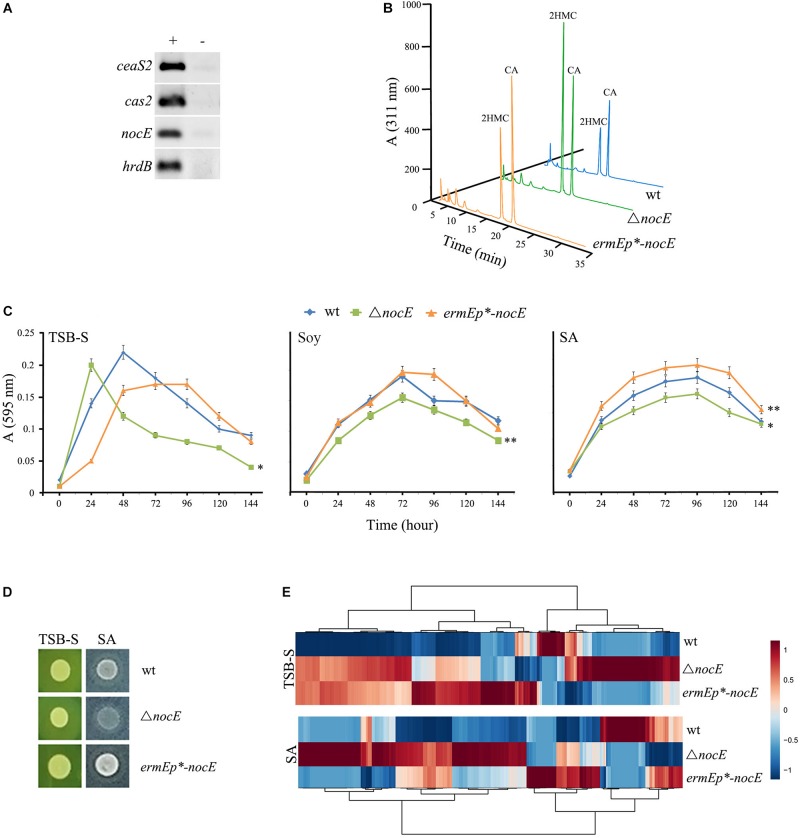
In previous studies, every gene from the proposed CA BGC of S. clavuligerus (Figure 3A) was systematically disrupted (Supplementary Table S1), to determine if it had any effect on CA or Ceph-C production. It has been suggested that nocE might have some role during CA biosynthesis in S. clavuligerus, but since the gene is not part of the CA BGC, a mutant has not been prepared and analyzed to date (Jensen, 2012). Therefore, the function of nocE was examined in the model CA producer, S. clavuligerus. RT-PCR analysis of RNA isolated from wt S. clavuligerus grown in SA medium demonstrated that nocE is temporally expressed along with ceaS2 and cas2 (Figure 4A), genes that are essential for CA biosynthesis (Figure 1). However, when S. clavuligerus strains were prepared in which nocE was either deleted (ΔnocE) or constitutively expressed (ermEp∗-nocE) (Table 1), the production of CA, 5S clavams, or Ceph-C was found to be unaffected (Figure 4B and Supplementary Figure S3), demonstrating that the gene is not required for β-lactam metabolite production in S. clavuligerus. The predicted lipase/esterase-like domain present in NocE is also found in hydrolytic enzymes from other Streptomyces species, some of which are known to be secreted (Wei et al., 1995; Vujaklija et al., 2002). Closer examination of the predicted NocE amino acid sequence from S. clavuligerus suggested that it is also a secreted protein, as it contains a highly conserved N-terminal Sec-signal sequence (p > 0.9) (Almagro Armenteros et al., 2019). These findings further ruled out the direct involvement of NocE in CA production, which occurs in the cytoplasm, and suggested that NocE might have some other exocellular hydrolytic function instead. Therefore, the S. clavuligerus wt, ΔnocE, and ermEp∗-nocE strains were assessed for growth under different nutritional condition using TSB-S (rich), soy (complex fermentation), or SA (defined fermentation) media (Figure 4C). It was observed that the growth of the S. clavuligerusΔnocE mutant was significantly reduced in each medium tested, whereas that of the ermEp∗-nocE strain was enhanced in SA medium only, when compared to the wt strain (Figure 4C). The growth of the three strains was also assessed on TSB-S and SA agar, which again showed that the S. clavuligerus ΔnocE mutant did not grow as well as the other strains in the latter medium (Figure 4D). To examine the influence of nocE on primary metabolism in S. clavuligerus, the wt, ΔnocE, and ermEp∗-nocE strains were grown on TSB-S and SA agar for metabolomics analysis, which showed marked differences in overall metabolite levels between the respective strains (Figure 4E). Furthermore, metabolomics analysis showed that SM production in S. clavuligerus was unaffected in the ΔnocE mutant as compared to the wt strain. Therefore, based on all evidence collected so far, it seems plausible that NocE could have some extracellular role in nutrient acquisition in S. clavuligerus, but like pcbR and orf11, it is not required for CA or Ceph-C production under the tested conditions.
FIGURE 4.
Examination of the function of nocE in S. clavuligerus. (A) RT-PCR analysis (+) of RNA isolated from 96-h S. clavuligerus SA cultures showing the expression of nocE during CA production. Transcription of ceaS2 and cas2 was used as a reporter for CA-BGC expression, whereas that of the constitutively expressed hrdB was used as a control. Negative controls (−) consisted of RNA samples subjected to PCR without undergoing RT. (B) LC-MS analysis of imidazole derivatized 96-h soy culture (different media from Figure 2B) supernatants form the S. clavuligerus wt, ΔnocE, and ermEp∗-nocE (constitutive expression) strains to assess CA and 5S clavam metabolite production. (C,D) Growth characteristics of the S. clavuligerus wt, ΔnocE, and ermEp∗-nocE strains in broth (D) or agar (D) cultures under different nutritional conditions, where (∗) and (∗∗) indicate p values of less than 0.05 and 0.001, respectively. (E) Comparative metabolomics of the S. clavuligerus wt, ΔnocE and ermEp∗-nocE strains grown on two different media as shown in panel (D). The heat map was constructed by hierarchical clustering of ∼1000 statically significant features to show overall differences between the three strains.
Conclusion
To summarize, we have shown that S. clavuligerus, S. jumonjinensis, and S. katsurahamanus contain numerous BGCs and that they synthesize many SMs, including the plant-associated metabolites, naringenin, and valerenic acid. It is possible that genes encoding enzymes for the synthesis of plant-associated metabolites are present in Streptomyces genomes, but they are not easily identified due to their organization, since some of them do not form BGCs (Álvarez-Álvarez et al., 2015; Nybo et al., 2017). In addition, plants normally produce metabolites like valerenic acid in low amounts, and for this reason, their heterologous production has been recently attempted in Saccharomyces and Escherichia coli (Nybo et al., 2017; Wong et al., 2018). The finding that certain Streptomyces species can synthesize these metabolites naturally could provide future avenues for their overproduction in a native host. Our results also show similarities and differences in the overall specialized metabolic capabilities of CA-producing Streptomyces species under different nutritional conditions, which, to the best of our knowledge, is the first report on the subject. Although the current study did not examine or address regulation, we would like to point out that many of the genes known to control Ceph-C and CA production in S. clavuligerus are also conserved in the two other producers (Liras et al., 2008; Ferguson et al., 2016; Fu et al., 2019b). It has been noted that deciphering the complete CA biosynthetic pathway in S. clavuligerus is challenging due to the presence of the 5S clavam biosynthetic pathway. The current report provides a framework for future studies on CA biosynthesis using S. jumonjinensis or S. katsurahamanus as models due to the absence of such competing or overlapping pathways in these organisms. Our analyses have also allowed us to propose the core group of genes involved in CA biosynthesis and have helped us to rule out the involvement of nocE and other genes in the production of this important metabolite.
Data Availability Statement
The datasets generated and/or analyzed during this study can be found in the NCBI sequence database (ncbi.nlm.nih.gov/genome) and the MassIVE public repository (massive.ucsd.edu). All accession numbers are provided in the Materials and Methods section.
Author Contributions
KT contributed conception, resources, and supervision. FB-G and PD provided reagents, resources, and supervision for genomics and metabolomics analysis, respectively. MM and PC-M performed the genome sequencing and annotation. NA and BP conducted the described comparative genomics analysis. NA prepared and analyzed the S. clavuligerus nocE mutant and overexpression strains. NA and SS prepared extracts for LC-MS/MS analysis, which was performed by L-FN. AS and L-FN carried out the metabolomics analysis and compound annotation. NA and MM wrote the first draft of the manuscript, whereas BP, AS, and L-FN wrote specific sections. NA, BP, AS, L-FN, FB-G, and KT contributed to manuscript revision.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Zhenglong Cheng (Memorial University of Newfoundland) for technical assistance. We would also like to express our deep gratitude to Dr. Susan E. Jensen (University of Alberta) for sharing the Streptomyces species used in the current study.
Funding. This work was supported by operating grants from the Natural Sciences and Engineering Research Council of Canada (NSERC: 386417–2010 and 2018–05949) to KT. NA and MM were the recipients of NSERC graduate student awards, including a Michael Smith Foreign Study Supplement to MM. Memorial University of Newfoundland also provided graduate student support to NA, BP, MM, and AS.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.02550/full#supplementary-material
References
- Abe I. (2018). Biosynthetic studies on teleocidins in Streptomyces. J. Antibiot. 71 763–768. 10.1038/s41429-018-0069-4 [DOI] [PubMed] [Google Scholar]
- Adusumilli R., Mallick P. (2017). Data conversion with proteowizard msconvert. Methods Mol. Biol. 1550 339–368. 10.1007/978-1-4939-6747-6_23 [DOI] [PubMed] [Google Scholar]
- Alexander D. C., Jensen S. E. (1998). Investigation of the Streptomyces clavuligerus cephamycin C gene cluster and its regulation by the CcaR protein. J. Bacteriol. 180 4068–4079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almagro Armenteros J. J., Tsirigos K. D., Sønderby C. K., Petersen T. N., Winther O., Brunak S., et al. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37 420–423. 10.1038/s41587-019-0036-z [DOI] [PubMed] [Google Scholar]
- Álvarez-Álvarez R., Botas A., Albillos S. M., Rumbero A., Martín J. F., Liras P. (2015). Molecular genetics of naringenin biosynthesis, a typical plant secondary metabolite produced by Streptomyces clavuligerus. Microb. Cell Fact. 14:178. 10.1186/s12934-015-0373-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez-Álvarez R., Martínez-Burgo Y., Pérez-Redondo R., Braña A. F., Martín J. F., Liras P. (2013). Expression of the endogenous and heterologous clavulanic acid cluster in Streptomyces flavogriseus: why a silent cluster is sleeping. Appl. Microbiol. Biotechnol. 97 9451–9463. 10.1007/s00253-013-5148-7 [DOI] [PubMed] [Google Scholar]
- Álvarez-Álvarez R., Martínez-Burgo Y., Rodríguez-García A., Liras P. (2017). Discovering the potential of S. clavuligerus for bioactive compound production: cross-talk between the chromosome and the pSCL4 megaplasmid. BMC Genomics 18:907. 10.1186/s12864-017-4289-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez-Álvarez R., Rodríguez-García A., Martínez-Burgo Y., Martín J. F., Liras P. (2018). Transcriptional studies on a Streptomyces clavuligerus oppA2 deletion mutant: n-acetylglycyl-clavaminic acid is an intermediate of clavulanic acid biosynthesis. Appl. Environ. Microbiol. 84:e1701-18. 10.1128/AEM.01701-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz R. H. (2008). Renaissance in antibacterial discovery from actinomycetes. Curr. Opin. Pharmacol. 8 557–563. 10.1016/j.coph.2008.04.008 [DOI] [PubMed] [Google Scholar]
- Barona-Gómez F., Wong U., Giannakopulos A. E., Derrick P. J., Challis G. L. (2004). Identification of a cluster of genes that directs desferrioxamine biosynthesis in Streptomyces coelicolor M145. J. Am. Chem. Soc. 126 16282–16283. 10.1021/ja045774k [DOI] [PubMed] [Google Scholar]
- Bentley S. D., Chater K. F., Cerdeno-Tarraga A. M., Challis G. L., Thomson N. R., James K. D., et al. (2002). Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417 141–147. 10.1038/417141a [DOI] [PubMed] [Google Scholar]
- Bibb M. J., Janssen G. R., Ward J. M. (1985). Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythraeus. Gene 38 215–226. 10.1016/0378-1119(85)90220-3 [DOI] [PubMed] [Google Scholar]
- Blin K., Wolf T., Chevrette M. G., Lu X., Schwalen C. J., Kautsar S. A., et al. (2017). antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 45 W36–W41. 10.1093/nar/gkx319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böcker S., Letzel M. C., Lipták Z., Pervukhin A. (2009). SIRIUS: decomposing isotope patterns for metabolite identification. Bioinformatics 25 218–224. 10.1093/bioinformatics/btn603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A. M., Lohse M., Usadel B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30 2114–2120. 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A. G. (1986). Clavulanic acid, a novel beta-lactamase inhibitor-a case study in drug discovery and development. Drug Des. Deliv. 1 1–21. [PubMed] [Google Scholar]
- Brown A. G., Butterworth D., Cole M., Hanscomb G., Hood J. D., Reading C., et al. (1976). Naturally-occurring beta-lactamase inhibitors with antibacterial activity. J. Antibiot. 29 668–669. 10.7164/antibiotics.29.668 [DOI] [PubMed] [Google Scholar]
- Brown D., Evans J. R., Fletton R. A. (1979). Structures of three novel β-lactams isolated from Streptomyces clavuligerus. J. Chem. Soc. Chem. Commun. 6 282–283. 10.1039/c39790000282 [DOI] [Google Scholar]
- Burrell M., Hanfrey C. C., Kinch L. N., Elliott K. A., Michael A. J. (2012). Evolution of a novel lysine decarboxylase in siderophore biosynthesis. Mol. Microbiol. 86 485–499. 10.1111/j.1365-2958.2012.08208.x [DOI] [PubMed] [Google Scholar]
- Cao G., Zhong C., Zong G., Fu J., Liu Z., Zhang G., et al. (2016). Complete genome sequence of Streptomyces clavuligerus F613-1, an industrial producer of clavulanic acid. Genome Announc. 4:e1020-16. 10.1128/genomeA.01020-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis G. L. (2005). A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. Chembiochem 6 601–611. 10.1002/cbic.200400283 [DOI] [PubMed] [Google Scholar]
- Challis G. L., Hopwood D. A. (2003). Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc. Natl. Acad. Sci. U.S.A. 100(Suppl. 2), 14555–14561. 10.1073/pnas.1934677100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W., Qu D., Zhai L., Tao M., Wang Y., Lin S., et al. (2010). Characterization of the tunicamycin gene cluster unveiling unique steps involved in its biosynthesis. Protein Cell 1 1093–1105. 10.1007/s13238-010-0127-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J., Soufan O., Li C., Caraus I., Li S., Bourque G., et al. (2018). MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 46 W486–W494. 10.1093/nar/gky310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech L., Hermann L., Stöveken N., Richter A. A., Hoppner A., Smits S. H. J., et al. (2018). Role of the extremolytes ectoine and hydroxyectoine as stress protectants and nutrients: genetics, phylogenomics, biochemistry, and structural analysis. Genes 9:E177. 10.3390/genes9040177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidsen J. M., Townsend C. A. (2009). Identification and characterization of NocR as a positive transcriptional regulator of the beta-lactam nocardicin A in Nocardia uniformis. J. Bacteriol. 191 1066–1077. 10.1128/JB.01833-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Fuente A., Lorenzana L. M., Martín J. F., Liras P. (2002). Mutants of Streptomyces clavuligerus with disruptions in different genes for clavulanic acid biosynthesis produce large amounts of holomycin: possible cross-regulation of two unrelated secondary metabolic pathways. J. Bacteriol. 184 6559–6565. 10.1128/jb.184.23.6559-6565.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillman R. O. (2004). Pentostatin (Nipent) in the treatment of chronic lymphocyte leukemia and hairy cell leukemia. Expert Rev. Anticancer Ther. 4 27–36. 10.1586/14737140.4.1.27 [DOI] [PubMed] [Google Scholar]
- Drawz S. M., Bonomo R. A. (2010). Three decades of beta-lactamase inhibitors. Clin. Microbiol. Rev. 23 160–201. 10.1128/CMR.00037-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan L. A., Busby R. W., Iwata-Reuyl D., Townsend C. A. (1997). Probable role of clavaminic acid as the terminal intermediate in the common pathway to clavulanic acid and the antipodal clavam metabolites. J. Am. Chem. Soc. 119 2348–2355. 10.1021/ja963107o [DOI] [Google Scholar]
- Ferguson N. L., Peña-Castillo L., Moore M. A., Bignell D. R., Tahlan K. (2016). Proteomics analysis of global regulatory cascades involved in clavulanic acid production and morphological development in Streptomyces clavuligerus. J. Ind. Microbiol. Biotechnol. 43 537–555. 10.1007/s10295-016-1733-y [DOI] [PubMed] [Google Scholar]
- Fernández-Martínez L. T., Bibb M. J. (2014). Use of the meganuclease I-SceI of Saccharomyces cerevisiae to select for gene deletions in actinomycetes. Sci. Rep. 4:7100. 10.1038/srep07100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Qin R., Zong G., Liu C., Kang N., Zhong C., et al. (2019a). The CagRS two-component system regulates clavulanic acid metabolism via multiple pathways in Streptomyces clavuligerus F613-1. Front. Microbiol. 10:244. 10.3389/fmicb.2019.00244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Qin R., Zong G., Zhong C., Zhang P., Kang N., et al. (2019b). The two-component system CepRS regulates the cephamycin C biosynthesis in Streptomyces clavuligerus F613-1. AMB Express 9:118. 10.1186/s13568-019-0844-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsior M., Mühlenweg A., Tietzmann M., Rausch S., Poch A., Süssmuth R. D. (2015). Biosynthesis of the peptide antibiotic feglymycin by a linear nonribosomal peptide synthetase mechanism. Chembiochem 16 2610–2614. 10.1002/cbic.201500432 [DOI] [PubMed] [Google Scholar]
- Gretes M., Lim D. C., de Castro L., Jensen S. E., Kang S. G., Lee K. J., et al. (2009). Insights into positive and negative requirements for protein-protein interactions by crystallographic analysis of the beta-lactamase inhibitory proteins BLIP, BLIP-I, and BLP. J. Mol. Biol. 389 289–305. 10.1016/j.jmb.2009.03.058 [DOI] [PubMed] [Google Scholar]
- Gunsior M., Breazeale S. D., Lind A. J., Ravel J., Janc J. W., Townsend C. A. (2004). The biosynthetic gene cluster for a monocyclic beta-lactam antibiotic, nocardicin A. Chem. Biol. 11 927–938. 10.1016/j.chembiol.2004.04.012 [DOI] [PubMed] [Google Scholar]
- Gurevich A., Saveliev V., Vyahhi N., Tesler G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29 1072–1075. 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamed R. B., Gomez-Castellanos J. R., Henry L., Ducho C., McDonough M. A., Schofield C. J. (2013). The enzymes of beta-lactam biosynthesis. Nat. Prod. Rep. 30 21–107. 10.1039/c2np20065a [DOI] [PubMed] [Google Scholar]
- Heikkila J., Akerman K. E. (1989). (-)-Indolactam V activates protein kinase C and induces changes in muscarinic receptor functions in SH-SY5Y human neuroblastoma cells. Biochem. Biophys. Res. Commun. 162 1207–1213. 10.1016/0006-291x(89)90802-4 [DOI] [PubMed] [Google Scholar]
- Hetrick K. J., van der Donk W. A. (2017). Ribosomally synthesized and post-translationally modified peptide natural product discovery in the genomic era. Curr. Opin. Chem. Biol. 38 36–44. 10.1016/j.cbpa.2017.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang S., Lee N., Jeong Y., Lee Y., Kim W., Cho S., et al. (2019). Primary transcriptome and translatome analysis determines transcriptional and translational regulatory elements encoded in the Streptomyces clavuligerus genome. Nucleic Acids Res. 47 6114–6129. 10.1093/nar/gkz471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen S. E. (2012). Biosynthesis of clavam metabolites. J. Ind. Microbiol. Biotechnol. 39 1407–1419. 10.1007/s10295-012-1191-0 [DOI] [PubMed] [Google Scholar]
- Jensen S. E., Elder K. J., Aidoo K. A., Paradkar A. S. (2000). Enzymes catalyzing the early steps of clavulanic acid biosynthesis are encoded by two sets of paralogous genes in Streptomyces clavuligerus. Antimicrob. Agents Chemother. 44 720–726. 10.1128/aac.44.3.720-726.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen S. E., Paradkar A. S. (1999). Biosynthesis and molecular genetics of clavulanic acid. Antonie Van Leeuwenhoek 75 125–133. [DOI] [PubMed] [Google Scholar]
- Jensen S. E., Paradkar A. S., Mosher R. H., Anders C., Beatty P. H., Brumlik M. J., et al. (2004a). Five additional genes are involved in clavulanic acid biosynthesis in Streptomyces clavuligerus. Antimicrob. Agents Chemother. 48 192–202. 10.1128/aac.48.1.192-202.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen S. E., Wong A., Griffin A., Barton B. (2004b). Streptomyces clavuligerus has a second copy of the proclavaminate amidinohydrolase gene. Antimicrob. Agents Chemother. 48 514–520. 10.1128/aac.48.2.514-520.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinovskaya N. I., Romanenko L. A., Irisawa T., Ermakova S. P., Kalinovsky A. I. (2011). Marine isolate Citricoccus sp. KMM 3890 as a source of a cyclic siderophore nocardamine with antitumor activity. Microbiol. Res. 166 654–661. 10.1016/j.micres.2011.01.004 [DOI] [PubMed] [Google Scholar]
- Kanno S., Tomizawa A., Hiura T., Osanai Y., Shouji A., Ujibe M., et al. (2005). Inhibitory effects of naringenin on tumor growth in human cancer cell lines and sarcoma S-180-implanted mice. Biol. Pharm. Bull. 28 527–530. 10.1248/bpb.28.527 [DOI] [PubMed] [Google Scholar]
- Katz L., Baltz R. H. (2016). Natural product discovery: past, present, and future. J. Ind. Microbiol. Biotechnol. 43 155–176. 10.1007/s10295-015-1723-5 [DOI] [PubMed] [Google Scholar]
- Kenig M., Reading C. (1979). Holomycin and an antibiotic (MM 19290) related to tunicamycin, metabolites of Streptomyces clavuligerus. J. Antibiot. 32 549–554. 10.7164/antibiotics.32.549 [DOI] [PubMed] [Google Scholar]
- Kieser T., Bibb M. J., Buttner M. J., Chater K. F., Hopwood D. A. (2000). Practical Streptomyces Genetics. Norwich: The John Innes Foundation. [Google Scholar]
- Kwong T., Tahlan K., Anders C. L., Jensen S. E. (2013). Carboxyethylarginine synthase genes show complex cross-regulation in Streptomyces clavuligerus. Appl. Environ. Microbiol. 79 240–249. 10.1128/AEM.02600-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Walsh C. T. (2010). Identification of the gene cluster for the dithiolopyrrolone antibiotic holomycin in Streptomyces clavuligerus. Proc. Natl. Acad. Sci. U.S.A. 107 19731–19735. 10.1073/pnas.1014140107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Khaleeli N., Townsend C. A. (2000). Expansion of the clavulanic acid gene cluster: identification and in vivo functional analysis of three new genes required for biosynthesis of clavulanic acid by Streptomyces clavuligerus. J. Bacteriol. 182 4087–4095. 10.1128/jb.182.14.4087-4095.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liras P. (1999). Biosynthesis and molecular genetics of cephamycins. Cephamycins produced by actinomycetes. Antonie Van Leeuwenhoek 75 109–124. [DOI] [PubMed] [Google Scholar]
- Liras P., Gomez-Escribano J. P., Santamarta I. (2008). Regulatory mechanisms controlling antibiotic production in Streptomyces clavuligerus. J. Ind. Microbiol. Biotechnol. 35 667–676. 10.1007/s10295-008-0351-8 [DOI] [PubMed] [Google Scholar]
- Liras P., Rodríguez-García A., Martín J. F. (1998). Evolution of the clusters of genes for beta-lactam antibiotics: a model for evolutive combinatorial assembly of new beta-lactams. Int. Microbiol. 1 271–278. [PubMed] [Google Scholar]
- Lucas X., Senger C., Erxleben A., Gruning B. A., Doring K., Mosch J., et al. (2013). StreptomeDB: a resource for natural compounds isolated from Streptomyces species. Nucleic Acids Res. 41 D1130–D1136. 10.1093/nar/gks1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Huang H., Liang J., Wang M., Lu L., Shao Z., et al. (2013). Activation and characterization of a cryptic polycyclic tetramate macrolactam biosynthetic gene cluster. Nat. Commun. 4:2894. 10.1038/ncomms3894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Burgo Y., Álvarez-Álvarez R., Rodríguez-García A., Liras P. (2015). The pathway-specific regulator ClaR of Streptomyces clavuligerus has a global effect on the expression of genes for secondary metabolism and differentiation. Appl. Environ. Microbiol. 81 6637–6648. 10.1128/AEM.00916-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Burgo Y., Santos-Aberturas J., Rodríguez-García A., Barreales E. G., Tormo J. R., Truman A. W., et al. (2019). Activation of secondary metabolite gene clusters in Streptomyces clavuligerus by the pimm regulator of Streptomyces natalensis. Front. Microbiol. 10:580. 10.3389/fmicb.2019.00580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema M. H., Trefzer A., Kovalchuk A., van den Berg M., Muller U., Heijne W., et al. (2010). The sequence of a 1.8-mb bacterial linear plasmid reveals a rich evolutionary reservoir of secondary metabolic pathways. Genome Biol. Evol. 2 212–224. 10.1093/gbe/evq013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellado E., Lorenzana L. M., Rodriguez-Saiz M., Diez B., Liras P., Barredo J. L. (2002). The clavulanic acid biosynthetic cluster of Streptomyces clavuligerus: genetic organization of the region upstream of the car gene. Microbiology 148(Pt 5), 1427–1438. 10.1099/00221287-148-5-1427 [DOI] [PubMed] [Google Scholar]
- Mosher R. H., Paradkar A. S., Anders C., Barton B., Jensen S. E. (1999). Genes specific for the biosynthesis of clavam metabolites antipodal to clavulanic acid are clustered with the gene for clavaminate synthase 1 in Streptomyces clavuligerus. Antimicrob. Agents Chemother. 43 1215–1224. 10.1128/aac.43.5.1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nybo S. E., Saunders J., McCormick S. P. (2017). Metabolic engineering of Escherichia coli for production of valerenadiene. J. Biotechnol. 262 60–66. 10.1016/j.jbiotec.2017.10.004 [DOI] [PubMed] [Google Scholar]
- Ohnishi Y., Ishikawa J., Hara H., Suzuki H., Ikenoya M., Ikeda H., et al. (2008). Genome sequence of the streptomycin-producing microorganism Streptomyces griseus IFO 13350. J. Bacteriol. 190 4050–4060. 10.1128/JB.00204-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivieri N. F., Brittenham G. M. (1997). Iron-chelating therapy and the treatment of thalassemia. Blood 89 739–761. 10.1182/blood.v89.3.739 [DOI] [PubMed] [Google Scholar]
- Onaka H., Taniguchi S., Igarashi Y., Furumai T. (2002). Cloning of the staurosporine biosynthetic gene cluster from Streptomyces sp. TP-A0274 and its heterologous expression in Streptomyces lividans. J. Antibiot. 55 1063–1071. 10.7164/antibiotics.55.1063 [DOI] [PubMed] [Google Scholar]
- Overbeek R., Olson R., Pusch G. D., Olsen G. J., Davis J. J., Disz T., et al. (2014). The seed and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42 D206–D214. 10.1093/nar/gkt1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradkar A. (2013). Clavulanic acid production by Streptomyces clavuligerus: biogenesis, regulation and strain improvement. J. Antibiot. 66 411–420. 10.1038/ja.2013.26 [DOI] [PubMed] [Google Scholar]
- Paradkar A. S., Aidoo K. A., Wong A., Jensen S. E. (1996). Molecular analysis of a beta-lactam resistance gene encoded within the cephamycin gene cluster of Streptomyces clavuligerus. J. Bacteriol. 178 6266–6274. 10.1128/jb.178.21.6266-6274.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradkar A. S., Jensen S. E. (1995). Functional analysis of the gene encoding the clavaminate synthase 2 isoenzyme involved in clavulanic acid biosynthesis in Streptomyces clavuligerus. J. Bacteriol. 177 1307–1314. 10.1128/jb.177.5.1307-1314.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Llarena F. J., Liras P., Rodríguez-García A., Martín J. F. (1997). A regulatory gene (ccaR) required for cephamycin and clavulanic acid production in Streptomyces clavuligerus: amplification results in overproduction of both beta-lactam compounds. J. Bacteriol. 179 2053–2059. 10.1128/jb.179.6.2053-2059.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluskal T., Castillo S., Villar-Briones A., Oresic M. (2010). MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11:395. 10.1186/1471-2105-11-395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prabhu J., Schauwecker F., Grammel N., Keller U., Bernhard M. (2004). Functional expression of the ectoine hydroxylase gene (thpD) from Streptomyces chrysomallus in Halomonas elongata. Appl. Environ. Microbiol. 70 3130–3132. 10.1128/aem.70.5.3130-3132.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruess D. L., Kellett M. (1983). Ro 22-5417, a new clavam antibiotic from Streptomyces clavuligerus. I. Discovery and biological activity. J. Antibiot. 36 208–212. 10.7164/antibiotics.36.208 [DOI] [PubMed] [Google Scholar]
- Rauha J. P., Remes S., Heinonen M., Hopia A., Kahkonen M., Kujala T., et al. (2000). Antimicrobial effects of Finnish plant extracts containing flavonoids and other phenolic compounds. Int. J. Food Microbiol. 56 3–12. [DOI] [PubMed] [Google Scholar]
- Romero J., Liras P., Martín J. F. (1984). Dissociation of cephamycin and clavulanic acid biosynthesis in Streptomyces clavuligerus. Appl. Microbiol. Biotechnol. 20 318–325. 10.1093/femsle/fnv215 [DOI] [PubMed] [Google Scholar]
- Romero J., Martín J. F., Liras P., Demain A. L., Rius N. (1997). Partial purification, characterization and nitrogen regulation of the lysine epsilon-aminotransferase of Streptomyces clavuligerus. J. Ind Microbiol. Biotechnol. 18 241–246. [DOI] [PubMed] [Google Scholar]
- Rudolf J. D., Yan X., Shen B. (2016). Genome neighborhood network reveals insights into enediyne biosynthesis and facilitates prediction and prioritization for discovery. J. Ind. Microbiol. Biotechnol. 43 261–276. 10.1007/s10295-015-1671-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M. A., et al. (2000). Artemis: sequence visualization and annotation. Bioinformatics 16 944–945. 10.1093/bioinformatics/16.10.944 [DOI] [PubMed] [Google Scholar]
- Sadeghi A., Soltani B. M., Nekouei M. K., Jouzani G. S., Mirzaei H. H., Sadeghizadeh M. (2014). Diversity of the ectoines biosynthesis genes in the salt tolerant Streptomyces and evidence for inductive effect of ectoines on their accumulation. Microbiol. Res. 169 699–708. 10.1016/j.micres.2014.02.005 [DOI] [PubMed] [Google Scholar]
- Sambrook J. R. D. (2001). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Saudagar P. S., Survase S. A., Singhal R. S. (2008). Clavulanic acid: a review. Biotechnol. Adv. 26 335–351. 10.1016/j.biotechadv.2008.03.002 [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., et al. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13 2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simao F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Skinnider M. A., Merwin N. J., Johnston C. W., Magarvey N. A. (2017). PRISM 3: expanded prediction of natural product chemical structures from microbial genomes. Nucleic Acids Res. 45 W49–W54. 10.1093/nar/gkx320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. Y., Jensen S. E., Lee K. J. (2010a). Clavulanic acid biosynthesis and genetic manipulation for its overproduction. Appl. Microbiol. Biotechnol. 88 659–669. 10.1007/s00253-010-2801-2 [DOI] [PubMed] [Google Scholar]
- Song J. Y., Jeong H., Yu D. S., Fischbach M. A., Park H. S., Kim J. J., et al. (2010b). Draft genome sequence of Streptomyces clavuligerus NRRL 3585, a producer of diverse secondary metabolites. J. Bacteriol. 192 6317–6318. 10.1128/JB.00859-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J. Y., Kim E. S., Kim D. W., Jensen S. E., Lee K. J. (2009). A gene located downstream of the clavulanic acid gene cluster in Streptomyces clavuligerus ATCC 27064 encodes a putative response regulator that affects clavulanic acid production. J. Ind. Microbiol. Biotechnol. 36 301–311. 10.1007/s10295-008-0499-2 [DOI] [PubMed] [Google Scholar]
- Spicer R. A., Salek R., Steinbeck C. (2017). Compliance with minimum information guidelines in public metabolomics repositories. Sci. Data 4:170137. 10.1038/sdata.2017.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava S. K., King K. S., AbuSara N. F., Malayny C. J., Piercey B. M., Wilson J. A., et al. (2019). In vivo functional analysis of a class A beta-lactamase-related protein essential for clavulanic acid biosynthesis in Streptomyces clavuligerus. PLoS One 14:e0215960. 10.1371/journal.pone.0215960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahlan K., Anders C., Wong A., Mosher R. H., Beatty P. H., Brumlik M. J., et al. (2007). 5S clavam biosynthetic genes are located in both the clavam and paralog gene clusters in Streptomyces clavuligerus. Chem. Biol. 14 131–142. 10.1016/j.chembiol.2006.11.012 [DOI] [PubMed] [Google Scholar]
- Tahlan K., Moore M. A., Jensen S. E. (2017). delta-(L-alpha-aminoadipyl)-L-cysteinyl-D-valine synthetase (ACVS): discovery and perspectives. J. Ind. Microbiol. Biotechnol. 44 517–524. 10.1007/s10295-016-1850-7 [DOI] [PubMed] [Google Scholar]
- Tahlan K., Park H. U., Jensen S. E. (2004a). Three unlinked gene clusters are involved in clavam metabolite biosynthesis in Streptomyces clavuligerus. Can. J. Microbiol. 50 803–810. 10.1139/w04-070 [DOI] [PubMed] [Google Scholar]
- Tahlan K., Park H. U., Wong A., Beatty P. H., Jensen S. E. (2004b). Two sets of paralogous genes encode the enzymes involved in the early stages of clavulanic acid and clavam metabolite biosynthesis in Streptomyces clavuligerus. Antimicrob. Agents Chemother. 48 930–939. 10.1128/aac.48.3.930-939.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai W., Paradkar A. S., Jensen S. E. (2001). Construction and analysis of ss-lactamase-inhibitory protein (BLIP) non-producer mutants of Streptomyces clavuligerus. Microbiology 147(Pt 2), 325–335. 10.1099/00221287-147-2-325 [DOI] [PubMed] [Google Scholar]
- Townsend C. A. (2002). New reactions in clavulanic acid biosynthesis. Curr. Opin. Chem. Biol. 6 583–589. [DOI] [PubMed] [Google Scholar]
- Tyers M., Wright G. D. (2019). Drug combinations: a strategy to extend the life of antibiotics in the 21st century. Nat. Rev. Microbiol. 17 141–155. 10.1038/s41579-018-0141-x [DOI] [PubMed] [Google Scholar]
- Upton C., Buckley J. T. (1995). A new family of lipolytic enzymes? Trends Biochem. Sci. 20 178–179. [DOI] [PubMed] [Google Scholar]
- Valegård K., Iqbal A., Kershaw N. J., Ivison D., Généreux C., Dubus A., et al. (2013). Structural and mechanistic studies of the orf12 gene product from the clavulanic acid biosynthesis pathway. Acta Crystallogr. D Biol. Crystallogr. 69(Pt 8), 1567–1579. 10.1107/S0907444913011013 [DOI] [PubMed] [Google Scholar]
- van der Heul H. U., Bilyk B. L., McDowall K. J., Seipke R. F., van Wezel G. P. (2018). Regulation of antibiotic production in Actinobacteria: new perspectives from the post-genomic era. Nat. Prod. Rep. 35 575–604. 10.1039/c8np00012c [DOI] [PubMed] [Google Scholar]
- Vujaklija D., Schröder W., Abramiæ M., Zou P., Lešc̀ič I., Franke P., et al. (2002). A novel streptomycete lipase: cloning, sequencing and high-level expression of the Streptomyces rimosus GDS(L)-lipase gene. Arch. Microbiol. 178 124–130. 10.1007/s00203-002-0430-6 [DOI] [PubMed] [Google Scholar]
- Wang L., Tahlan K., Kaziuk T. L., Alexander D. C., Jensen S. E. (2004). Transcriptional and translational analysis of the ccaR gene from Streptomyces clavuligerus. Microbiology 150(Pt 12), 4137–4145. 10.1099/mic.0.27245-0 [DOI] [PubMed] [Google Scholar]
- Wang M., Carver J. J., Phelan V. V., Sanchez L. M., Garg N., Peng Y., et al. (2016). Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 34 828–837. 10.1038/nbt.3597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. M., Hodgson J. E. (1993). The biosynthetic genes for clavulanic acid and cephamycin production occur as a ‘super-cluster’ in three Streptomyces. FEMS Microbiol. Lett. 110 239–242. 10.1111/j.1574-6968.1993.tb06326.x [DOI] [PubMed] [Google Scholar]
- Wei Y., Schottel J. L., Derewenda U., Swenson L., Patkar S., Derewenda Z. S. (1995). A novel variant of the catalytic triad in the Streptomyces scabies esterase. Nat. Struct. Biol. 2 218–223. [DOI] [PubMed] [Google Scholar]
- Wong J., d’Espaux L., Dev I., van der Horst C., Keasling J. (2018). De novo synthesis of the sedative valerenic acid in Saccharomyces cerevisiae. Metab. Eng. 47 94–101. 10.1016/j.ymben.2018.03.005 [DOI] [PubMed] [Google Scholar]
- Wu P., Wan D., Xu G., Wang G., Ma H., Wang T., et al. (2017). An unusual protector-protege strategy for the biosynthesis of purine nucleoside antibiotics. Cell Chem. Biol. 24 171–181. 10.1016/j.chembiol.2016.12.012 [DOI] [PubMed] [Google Scholar]
- Yamanaka K., Oikawa H., Ogawa H. O., Hosono K., Shinmachi F., Takano H., et al. (2005). Desferrioxamine E produced by Streptomyces griseus stimulates growth and development of Streptomyces tanashiensis. Microbiology 151(Pt 9), 2899–2905. 10.1099/mic.0.28139-0 [DOI] [PubMed] [Google Scholar]
- Zelyas N. J., Cai H., Kwong T., Jensen S. E. (2008). Alanylclavam biosynthetic genes are clustered together with one group of clavulanic acid biosynthetic genes in Streptomyces clavuligerus. J. Bacteriol. 190 7957–7965. 10.1128/JB.00698-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino D. R., Birney E. (2008). Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 18 821–829. 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Zhang W., Saha S., Zhang C. (2016). Recent advances in discovery, biosynthesis and genome mining of medicinally relevant polycyclic tetramate macrolactams. Curr. Top. Med. Chem. 16 1727–1739. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Xiang S., Dai X., Yang K. (2013). A simplified diphenylamine colorimetric method for growth quantification. Appl. Microbiol. Biotechnol. 97 5069–5077. 10.1007/s00253-013-4893-y [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during this study can be found in the NCBI sequence database (ncbi.nlm.nih.gov/genome) and the MassIVE public repository (massive.ucsd.edu). All accession numbers are provided in the Materials and Methods section.