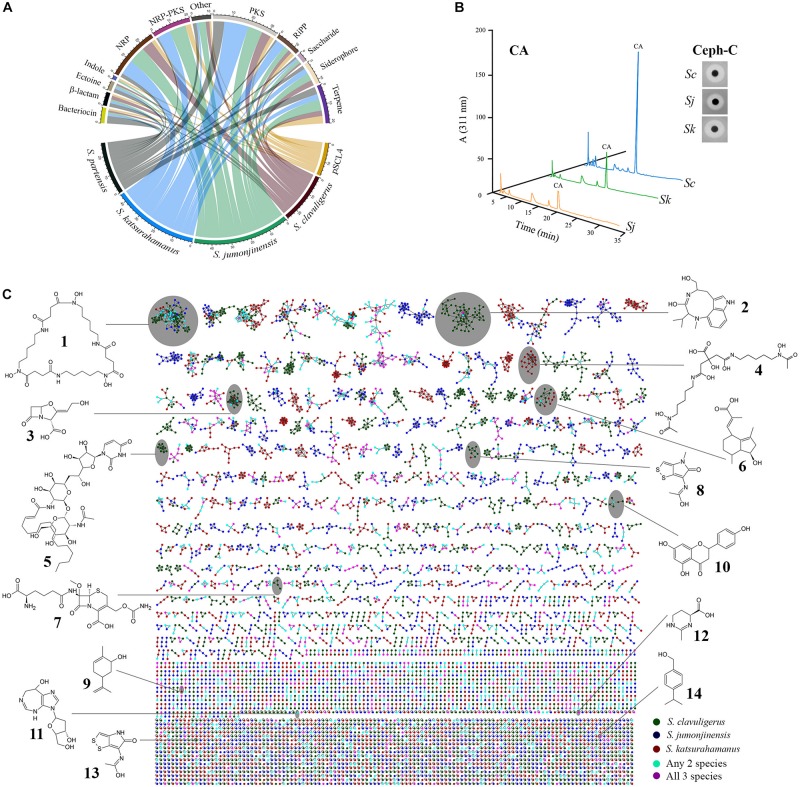
FIGURE 2.
Biosynthetic gene cluster (BGC) content and metabolomics analysis of clavulanic acid (CA)-producing Streptomyces species (S. clavuligerus, S. jumonjinensis, and S. katsurahamanus). (A) Circular chord diagram representing all predicted BGC classes present in the respective Streptomyces species. S. pratensis was included for comparison as the bacterium contains a CA-like BGC, but does not produce the metabolite. The sequence of the giant linear plasmid pSCL4 from S. clavuligerus was also included separately due to the presence of multiple BGCs on it. The lower arc represents genomes/plasmid, while the upper arc represents different classes of BGCs and the color-coded ribbons connecting them indicate the presence of a BGC in the specific species. (B) Detection of CA and cephamycin C (Ceph-C) in 96-h SA culture supernatants of S. clavuligerus (Sc), S. jumonjinensis (Sj), and S. katsurahamanus (Sk) using LC-MS (after imidazole derivatization) and bioassays (inset), respectively. The peak corresponding to CA in HPLC chromatograms is noted and the zones of inhibition in the inset panel demonstrate relative amounts of Ceph-C production. (C) Metabolic network constructed using S. clavuligerus, S. jumonjinensis, and S. katsurahamanus culture extracts (culture conditions and details are described in the section “Materials and Methods”). The network is color-coded according to source organism (bottom right legend), where each node depicts a mass spectrum and edges represent the relationship between different nodes. Structures of natural products detected in the extracts at high confidence in the three species are shown and the clade in the network containing the node corresponding to the respective metabolite is also indicated. 1, desferrioxamine E; 2, (−)-indolactam V; 3, clavulanic acid; 4, arthrobactin; 5, tunicamycin C2; 6, hydroxyvalerenic acid; 7, cephamycin C; 8, thiolutin; 9, (−)-carveol; 10, naringenin; 11, pentostatin, 12, ectoine; 13, holomycin; and 14, cuminyl alcohol.