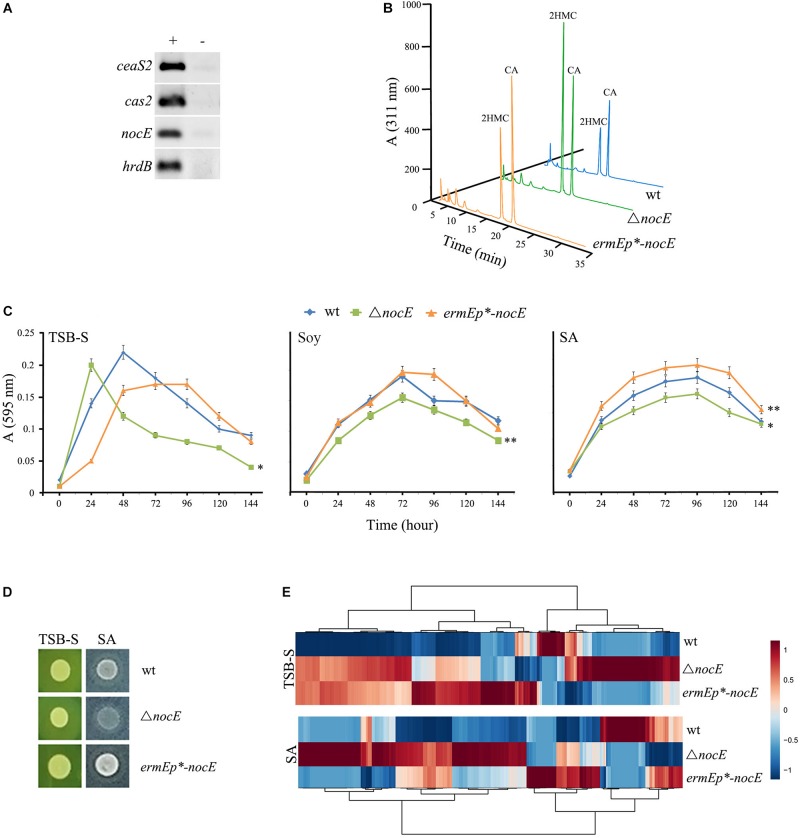
FIGURE 4.
Examination of the function of nocE in S. clavuligerus. (A) RT-PCR analysis (+) of RNA isolated from 96-h S. clavuligerus SA cultures showing the expression of nocE during CA production. Transcription of ceaS2 and cas2 was used as a reporter for CA-BGC expression, whereas that of the constitutively expressed hrdB was used as a control. Negative controls (−) consisted of RNA samples subjected to PCR without undergoing RT. (B) LC-MS analysis of imidazole derivatized 96-h soy culture (different media from Figure 2B) supernatants form the S. clavuligerus wt, ΔnocE, and ermEp∗-nocE (constitutive expression) strains to assess CA and 5S clavam metabolite production. (C,D) Growth characteristics of the S. clavuligerus wt, ΔnocE, and ermEp∗-nocE strains in broth (D) or agar (D) cultures under different nutritional conditions, where (∗) and (∗∗) indicate p values of less than 0.05 and 0.001, respectively. (E) Comparative metabolomics of the S. clavuligerus wt, ΔnocE and ermEp∗-nocE strains grown on two different media as shown in panel (D). The heat map was constructed by hierarchical clustering of ∼1000 statically significant features to show overall differences between the three strains.