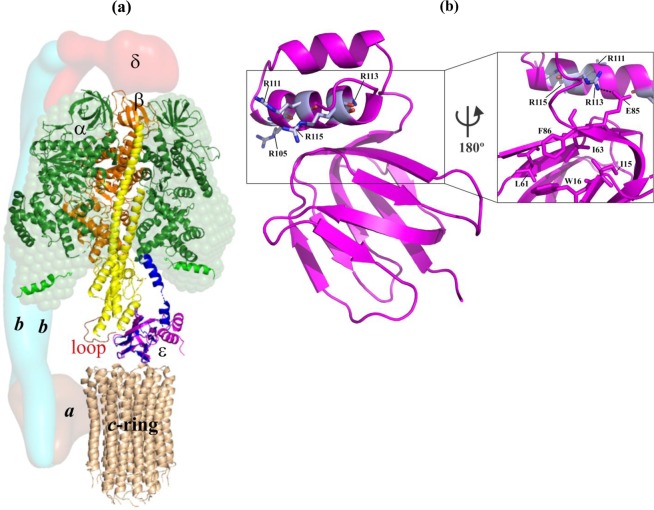
Figure 1.
Structural features of the F-ATP synthase and the mycobacterial coupling subunit ε. (a) The structural model of the mycobacterial F-ATP synthase was generated based on the cryo-EM model of E. coli F-ATP synthase (PDB ID: 5T4O)47 and the structural model of the α3β3γε complex of the mycobacterial F-ATP synthase7. Subunits α (green), β (orange), γ (yellow), and ε (magenta) are from the M. smegmatis crystal structure (PDB ID: 6FOC)7, whereby the missing β-sheet elements of subunit γ were added by the respective subunit γ elements of the E. coli F-ATP synthase (PDB ID: 5T4Q). Mycobacterial subunit γ has a unique γ-loop13 which is highlighted in red. The solution shape of αchi is displayed as green sphere14. The c-ring loop residues of M. phlei (wheat; PDB ID: 4V1G9), proposed to interact with the rotating ε and γ stalks subunits are indicated. The modelled extended ε subunit (blue) of M. tuberculosis (PDB ID: 5YIO) reaches the DELSEED-region in subunit α10. The low-resolution shapes of the related E. coli subunits a, b-dimer and δ are shown in the brown, light blue and red, respectively, and based on the EM map (PDB ID: 5T4O). (b) NMR solution structure of Mtε (PDB ID: 5YIO)10 showing R105A, R111A, R113A, and R115A (magenta) in helix-2 of the C-terminal domain. (Inset) Closer view of the back surface revealing the interaction of R113 with E85 of the N-terminal domain, which is in close proximity to the hydrophobic cleft proposed to bind BDQ. We thank Dr. S. S. M. Malathy for the art work of (a).