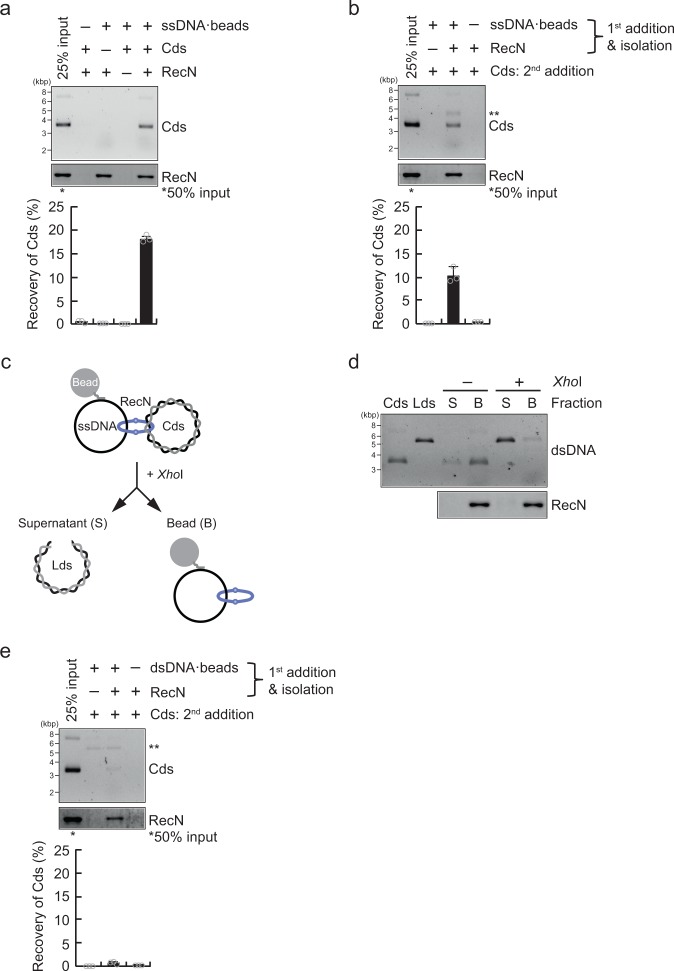
Fig. 2.
The ssDNA–dsDNA tethering activity of RecN via topological DNA binding. a One-step pull-down assays in which RecN and phiX174 circular dsDNA (Cds) were incubated at 37 °C for 10 min in the presence of DNA-free beads or ssDNA beads. The materials bound to the beads were eluted with SDS–sample buffer. The eluted proteins were analyzed by SDS–PAGE and CBB staining, and the eluted Cds was analyzed by agarose gel electrophoresis and SYBR Gold staining. The lower graph shows the intensities of the Cds bands in the agarose gel image. b Two-step pull-down assays in which RecN was incubated at 37 °C for 10 min in the presence of DNA-free beads or ssDNA beads. The beads were then washed and incubated with Cds as a second substrate. The bead-bound materials were eluted with SDS–sample buffer and analyzed as described above. The lower graph shows the intensities of the Cds bands in the agarose gel image. **ssDNA dissociated from the beads. c Schematic illustration of linear dsDNA (Lds) release by linearization of the captured second Cds in d. d The ternary complex comprising RecN, dsDNA, and ssDNA beads, isolated by the same method as described for b was treated with or without XhoI. The bead (B) and the supernatant (S) fractions were analyzed by agarose gel electrophoresis and SDS–PAGE as described above. Cds and Lds are shown as molecular markers. e Two-step pull-down assays using dsDNA beads, in which RecN was incubated at 37 °C for 10 min in the presence of DNA-free beads or dsDNA beads. The beads were then washed and incubated with Cds as a secondary substrate. The bead-bound materials were analyzed as described above. The lower graph shows the intensities of Cds bands in the agarose gel image. **dsDNA dissociated from the beads. a, b and e Data represent the mean ± standard deviation of three independent experiments