Fig. 3.
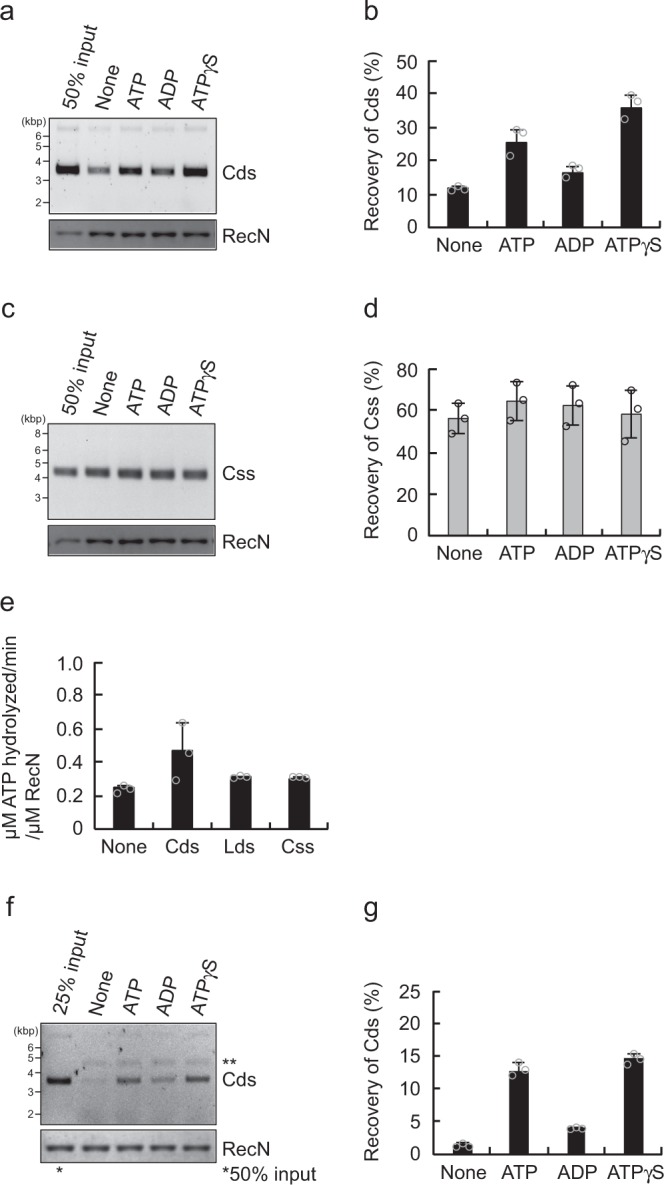
The effect of nucleotides on the DNA-binding activity of RecN. a RecN (0.5 µM) was incubated at 37 °C for 10 min in the presence of phiX174 circular dsDNA (Cds) and the indicated nucleotides (2 mM). The RecN–DNA complex was collected using Co2+-conjugated beads, washed in buffer containing 500 mM KCl, and eluted with SDS–sample buffer. The eluted proteins and DNA were analyzed as described for Fig. 1c. b Quantification of the intensities of the Cds bands in the agarose gel image shown in a. c The assay was performed as described for a, with the exception that phiX174 circular ssDNA (Css) was used in place of Cds. d Quantification of the intensities of the Css bands in the agarose gel image shown in c. e The ATPase activity of RecN. RecN (1 µM) was incubated at 37 °C for 60 min in reaction buffer containing 2 mM ATP plus [γ32P]-ATP, in the absence or presence of the indicated phiX174-derivative DNA substrates. The ATPase rates (µM ATP hydrolyzed min−1 per µM RecN) were calculated following quantification of the spots of radioactive inorganic phosphate (32Pi) using an image analyzer. f Two-step pull-down assays in the absence or presence of the indicated nucleotides (2 mM). Cds and RecN recovered by the ssDNA beads were analyzed as described for Fig. 2b. **ssDNA dissociated from the beads. g Quantification of the intensities of the Cds bands in the agarose gel image shown in f. b, d, e, and g All graphs show the mean ± standard deviation of three independent experiments
