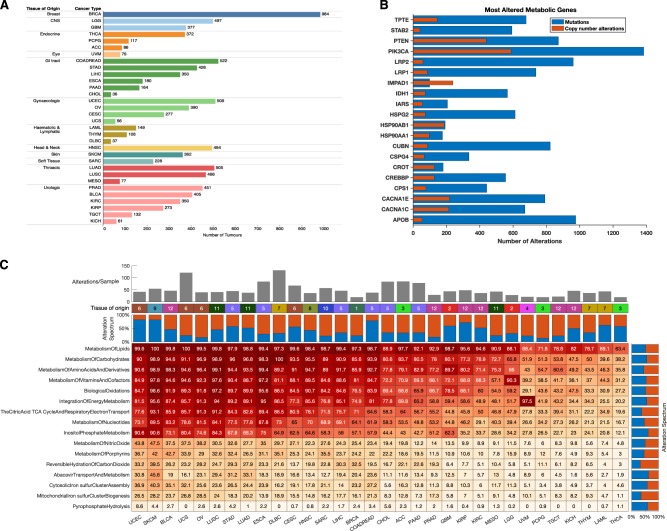
Fig. 1.
a Distribution of 10,528 TCGA tumours across 32 human cancer types broken down by tissue of origin. TCGA disease codes and abbreviations: UCEC, uterine corpus endometrial carcinoma; SKCM, skin cutaneous melanoma; BLCA, bladder urothelial carcinoma; UCS, uterine carcinosarcoma; OV, ovarian serous cystadenocarcinoma; LUSC, lung squamous cell carcinoma; STAD, stomach adenocarcinoma; LUAD, lung adenocarcinoma; ESCA, oesophageal adenocarcinoma; DLBC, diffuse large b-cell lymphoma; CESC, cervical squamous cell carcinoma; HNSC, head and neck squamous cell carcinoma; SARC, sarcoma; LIHC, liver hepatocellular carcinoma; BRCA, breast invasive carcinoma; COADREAD, colorectal adenocarcinoma; CHOL, cholangiocarcinoma; ACC, adrenocortical carcinoma; PAAD, pancreatic adenocarcinoma; PRAD, prostate adenocarcinoma; GBM, glioblastoma multiforme; KIRP, kidney renal papillary cell carcinoma; KIRC, kidney renal clear cell carcinoma; MESO, mesothelioma; LGG, brain lower grade glioma; UVM, uveal melanoma; PCPG, pheochromocytoma and paraganglioma; TGCT, testicular germ cell tumours; KICH, kidney chromophobe; THYM, thymoma; LAML, acute myeloid leukaemia; THCA, thyroid carcinoma. b Genes involved in metabolism found to be most altered across all human cancers. c Clustered heatmap of cancer types using the percentage of tumours with first-tier metabolic pathway genes displaying alterations. Pathways are ordered by decreasing frequencies of alterations. Increasing colour intensities denote higher percentages. The heat map was produced using unsupervised hierarchical clustering with the Euclidean distance metric and complete linkage (see Supplementary Fig. 1). The coloured bars on the heatmap show the tissue of origin for each cancer: 1 = Breast; 2 = CNS, 3 = Endocrine; 4 = Eye; 5 = GI tract; 6 = Gynaecologic; 7 = Haematologic & Lymphatic; 8 = Head & Neck; 9 = Skin; 10 = Soft Tissue; 11 = Thoracic; 12 = Urologic. The bar graph represents the overall frequency of genomic alterations in each human cancer