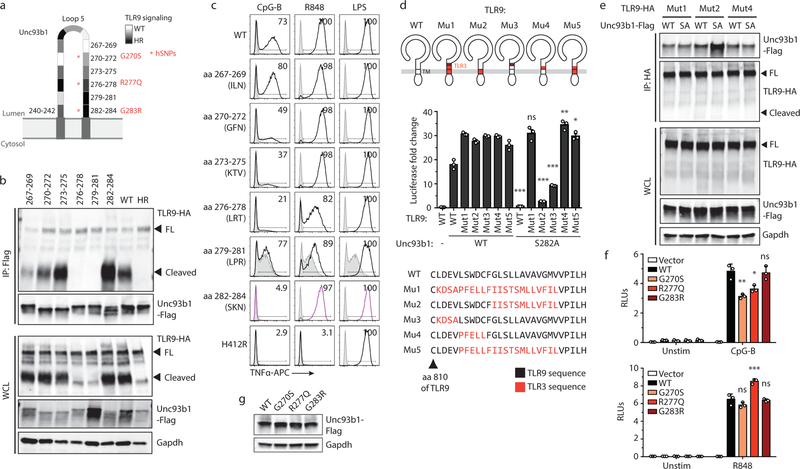
Extended Data Fig. 5: Identification of residues within loop 5 of Unc93b1 that mediate interaction with TLR9.
(a) Schematic of the tested loop 5 mutants of Unc93b1 and the relative TLR9 responses indicated in shades of gray: white indicates a response equivalent to WT while black indicates no response. Asterisks show human Unc93b1 SNPs that have been tested in (f). (b) A larger region in loop 5 of Unc93b1 mediates binding to TLR9. Immunoprecipitation of Unc93b1-Flag from macrophage lines expressing the indicated Unc93b1 mutants (spanning amino acids 267–284, and non-functional HR) followed by immunoblot of TLR9-HA. Representative of two independent experiments. (c) Intracellular cytokine staining of TNFα in macrophage lines shown in (b) after stimulation with CpG-B (25 nM), R848 (100 ng/ml), and LPS (10 ng/ml). Shaded histograms show unstimulated controls. Representative of three independent experiments. (d) Schematics showing relative positions and a sequence alignment (bottom) of swapped regions within the TLR9/3 chimeras. Colored regions indicate TLR3 sequences. NF-κB luciferase assay in HEK293T cells transiently transfected with the indicated TLR9 and Unc93b1 mutants and stimulated with CpG-B (200nM) for 16h. Data are normalized to Unc93b1-independent hIL-1b responses and expressed as luciferase fold change over unstimulated controls. Data are mean ± s.d., n=3 biological replicates. p-values determined by two-way ANOVA followed by a Sidak’s posttest comparing each TLR9 allele coexpressed with Unc93b1 WT vs S282A (TLR9WT: p<0.0001, TLR9Mut1: p>0.9999, TLR9Mut2: p<0.0001, TLR9Mut3: p<0.0001, TLR9Mut4: p=0.0020, TLR9Mut5: p=0.0171. Representative experiment of two independent repeats. (e) TLR9 mutants that rescue signaling in the presence of Unc93b1S282A also show normal binding to Unc93b1S282A. HA immunoprecipitation of the indicated TLR9 mutants transiently expressed in HEK293T cells that stably express the indicated Unc93b1-Flag alleles, followed by immunoblot of Unc93b1-Flag. Representative of three independent experiments. (f) Human Unc93b1 variants with SNPs in loop 5 show decreased TLR9 signaling. NF-κB luciferase assay in HEK293T cells expressing TLR9 or TLR7 and the indicated human Unc93b1-Flag variants and stimulated with CpG-B (250 nM) or R848 (250 ng/ml) for 16h, respectively. Data are normalized to Renilla expression and expressed as relative luciferase units (RLUs). Data are mean ± s.d., n=3 biological replicates. p-values are determined by one-way ANOVA followed by a Tukey’s posttest. For CpG-B stimulations: p=0.0048 (WT vs G270S), p=0.0113 (WT vs R277Q), p=0.9994 (WT vs G283R). Representative of four independent experiments. For R848 stimulations: p=0.2001 (WT vs G270S), p=0.0002 (WT vs R277Q), p=0.9933 (WT vs G283R). Representative of three independent repeats experiment. (g) Expression levels of the Unc93b1 mutants used in (f). *p< 0.05, **p< 0.01, ***p< 0.001, ns: not significant. WCL: whole cell lysate. FL: full-length.