Figure 1.
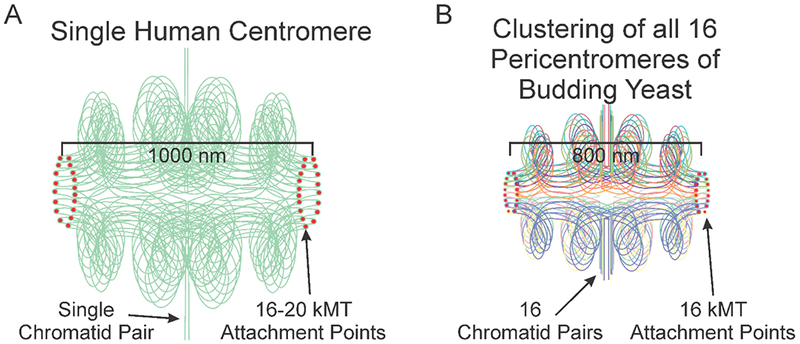
Depictions of a single human centromere and yeast pericentromere. A) A schematic of 2 sister chromatids at the centromere. One pair of sisters are aligned along the vertical axis. Within the centromere multiple loops are folded intramolecularly and biorient relative to one another. The apex of a loop is positioned at the kinetochore plus-ends (red dote). The sister chromatids are in green, highlighting the proposal that 16-20 loops emanate from one pair of sister chromatids in mammals (or organisms with multiple microtubule attachment sites) (Aze et al., 2016; Bloom and Costanzo, 2017), versus the clustering of loops from individual chromosomes (16 in budding yeast) in organisms with unit attachment sites. B) A schematic of the 16 sister pericentromeres in budding yeast. Pairs of sister chromatids are aligned along the vertical axis (vertical straight lines). In the pericentromere (horizontal, bounded by red kinetochore attachment sites), each sister centromere is folded into an intramolecular C-loop (Yeh et al., 2008) that biorient relative to one another that contains several subloops (Stephens et al., 2011). The apex of each C-loop (the 125 bp centromere) is positioned at the kinetochore microtubule plus-ends (red dot). The sister chromatids are color-coded (evident in the colors at the kinetochore microtubule attachment site). The schematics scaled by centromere separation, i.e. the cluster of 16 bi-oriented kinetochores in yeast occupy roughly the same volume as one mammalian kinetochore.
