Table 1.
Scaling of sister kinetochore separation across phylogeny. Kinetochore separation is the distance between sister kinetochores observed in a number of organisms O. tauri (Gan, Ladinsky, and Jensen, 2011); budding yeast, S. cerevisiae (Lawrimore et al., 2016; Yeh et al., 2008); worm, C. albicans (Maddox et al., 2006); fission yeast, S. pombe (Ding, McDonald, and McIntosh, 1993); fly, D. melanogaster (Venkei et al., 2012); and human, H. sapiens (Salmon et al., 1976)]. Centromere DNA size is defined as the region of DNA required for segregation function or physically located at the primary constriction in condensed mitotic chromosomes. Centromere DNA spans 4-5 orders of magnitude in size from yeast to human, while sister kinetochore separation scales by a factor of 2, from the smallest single-cell eukaryote to multicellular organisms.
| Centromere DNA size | Kinetochore separation in mitosis | |
|---|---|---|
| O. Tauri | ~0.1 kb | 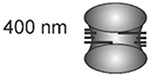 |
| S. cerevisiae | 0.125 kb | 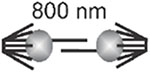 |
| C. albicans | 3-4 kb | ~800 nm |
| S. pombe | 10 kb | ~1,000 nm |
| D. melanogaster | 200-500 kb | ~1,000 nm |
| H. sapiens | 500-1,500 kb | 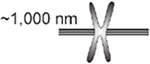 |
