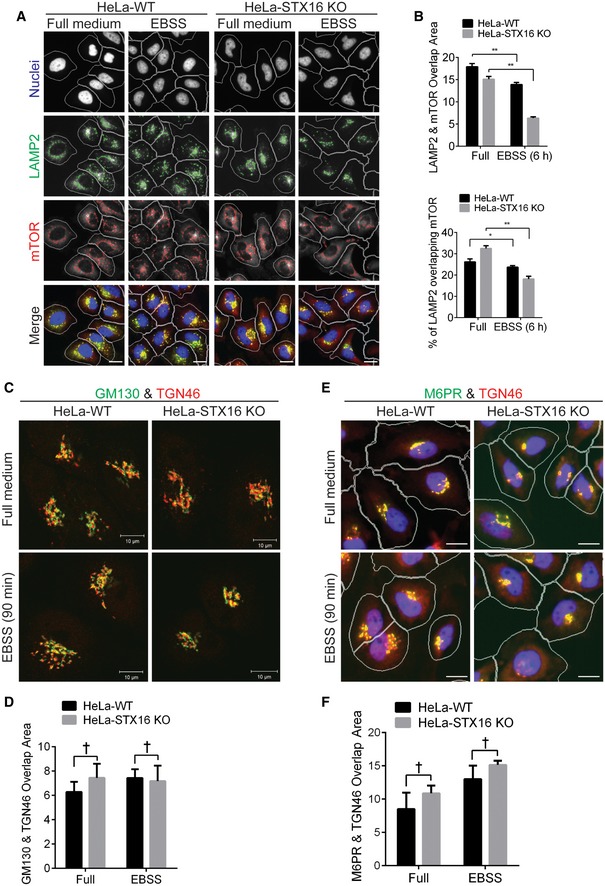
WT or STX16
KO HeLa cells were starved in EBSS for 6 h and subjected to HCM analysis of the overlap between LAMP2 and mTOR. Masks: white, cells identified based on nuclei; green, LAMP2 puncta; red, mTOR puncta; yellow, overlap between LAMP2 and mTOR. Scale bar: 20 μm.
Quantifications of overlaps between LAMP2 and mTOR in WT and
STX16
KO HeLa cells treated as in (D). Data shown as means ± SEM of overlap area per cell between LAMP2 and mTOR (upper panel), and percentages of LAMP2 overlapping with mTOR to compensate reduced LAMP2 puncta in
STX16
KO cells (see Fig
5D), minimum 500 cells were counted each well from at least 12 wells, three independent experiments; *
P < 0.05; **
P < 0.01 (two‐way ANOVA).
WT or STX16
KO HeLa cells were starved in EBSS for 90 min, fixed, and stained with GM130 and TGN46 antibodies, followed by confocal microscopy analysis of the GM130 (Golgi marker, green) and TGN46 (TGN marker, red) profiles. Scale bar, 10 μm.
HCM quantifications of overlaps between GM130 and TGN46 in WT and STX16
KO HeLa cells treated as in (A). Data shown as means ± SEM of overlap area per cell between GM130 and TGN46, minimum 500 cells were counted each well from at least 12 wells, three independent experiments; †, not significant (two‐way ANOVA).
WT or STX16
KO HeLa cells were starved in EBSS for 90 min, fixed, and stained with M6PR and TGN46 antibodies, followed by HCM analysis of the M6PR and TGN46 profiles. Masks: white, cells identified based on nuclei; green, M6PR puncta; red, TGN46; yellow, overlap between M6PR and TGN46. Scale bar: 20 μm.
HCM quantifications of overlaps between M6PR and TGN46 in WT and STX16
KO HeLa cells treated as in (C). Data shown as means ± SEM of overlap area per cell between M6PR and TGN46, minimum 500 cells were counted each well from at least 12 wells, three independent experiments; †, not significant (two‐way ANOVA).