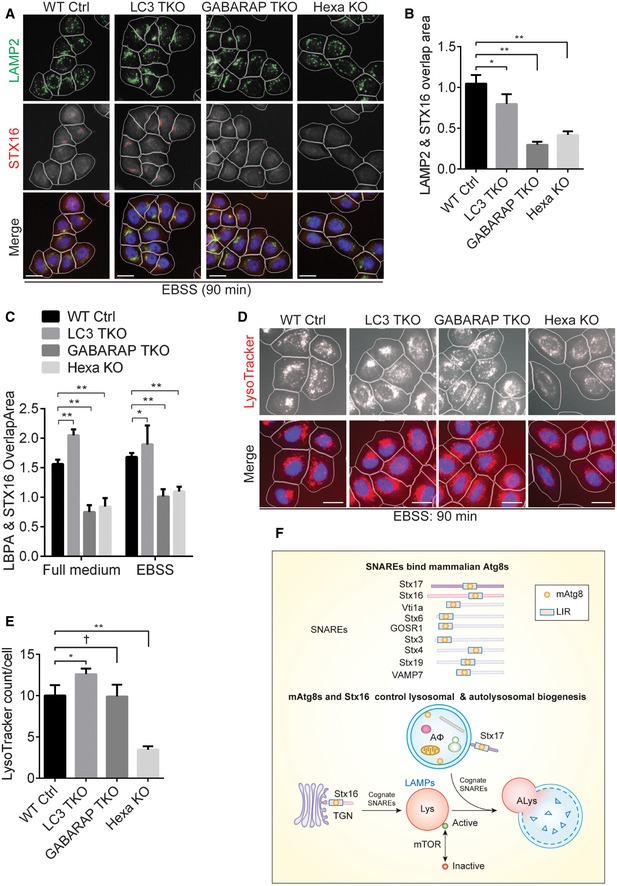
WT, LC3TKO, GABARAPTKO, or HexaKO HeLa cells were starved in EBSS for 90 min and subjected to HCM analysis of overlaps between LAMP2 and Stx16. Masks: white, cells identified based on nuclei; green, LAMP2 puncta; red, Stx16 puncta; yellow, overlap of LAMP2 and Stx16. Scale bar: 20 μm.
Quantifications of overlaps between LAMP2 and Stx16 in WT, LC3TKO, GABARAPTKO, or HexaKO HeLa cells treated as in (C). Data shown as means ± SEM of LAMP2 and Stx16 overlap area per cell, minimum 500 cells were counted each well from at least 12 wells, three independent experiments; *P < 0.05; **P < 0.01 (one‐way ANOVA).
Quantifications of overlaps between LBPA and Stx16 in WT, LC3
TKO, GABARAP
TKO, or Hexa
KO HeLa cells grown in full medium or treated as in (A). Data shown as means ± SEM of LBPA and Stx16 overlap area per cell, minimum 500 cells were counted each well from at least 12 wells, three independent experiments; *
P < 0.05; **
P < 0.01 (one‐way ANOVA). Representative images are shown in
Appendix Fig S5A.
WT, LC3TKO, GABARAPTKO, or HexaKO HeLa cells were starved in EBSS for 1 h, followed by starvation with the presence of LTR (100 nM) for additional 30 min, and subjected to HCM analysis of LTR. Scale bar: 20 μm.
Quantifications of LTR puncta in WT, LC3TKO, GABARAPTKO, or HexaKO HeLa cells treated as in (D). Data shown as means ± SEM of LTR puncta per cell, minimum 500 cells were counted each well from at least 12 wells, three independent experiments; †, not significant; *P < 0.05; **P < 0.01 (one‐way ANOVA).
Schematic model of this study. Mammalian Atg8 proteins directly bind Stx16 through its LIR motif, which controls the proper localization of the Stx16 SNARE complex at the endolysosomal compartments. The Stx16 SNARE complex functions in the maintenance of lysosomal homeostasis upon autophagy induction, thus controlling mTOR activity.