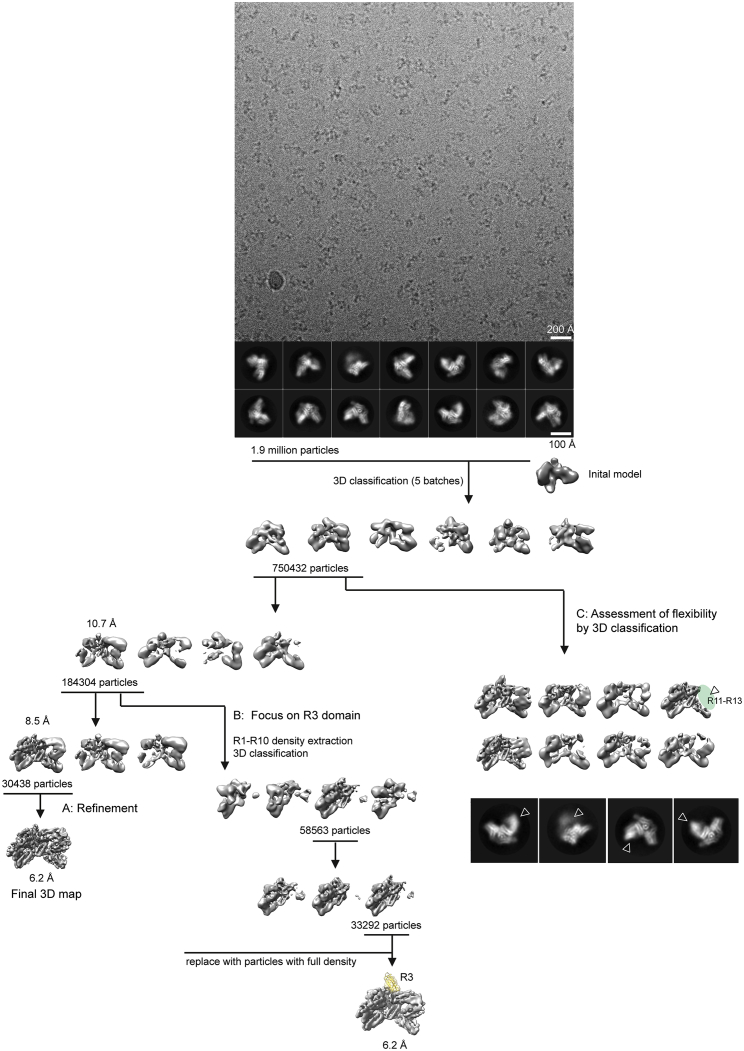
Figure S1.
Summary of Cryo-EM Data Processing, Related to Figure 1
Top: Representative cryo-EM image of talin-FL at 75 mM salt with representative 2D averages. Bottom: Graphical scheme of image processing. 1.9 million particles were selected out of 2D classifications. Further 3D classifications and refinement facilitated the final map generation (A – left). To visualize the R3 domain, the core density (R1-R10) was computationally extracted and focused 3D classification of the core density was performed. At the final step of the processing, the alignment parameters were applied to the original particles to visualize the full density (B – center). Yellow density fitted with an available structural model of R3. To assess the flexibility of the autoinhibition pocket consisting of the interface between FERM domain F2-F3 and R12-R13 (lid), extensive 3D classifications were performed, yielding a visualization of the flexible densities of R11-R13 (C – right).