Figure 6.
PRG2 Is Required for Filopodia and Branch Formation in Axons
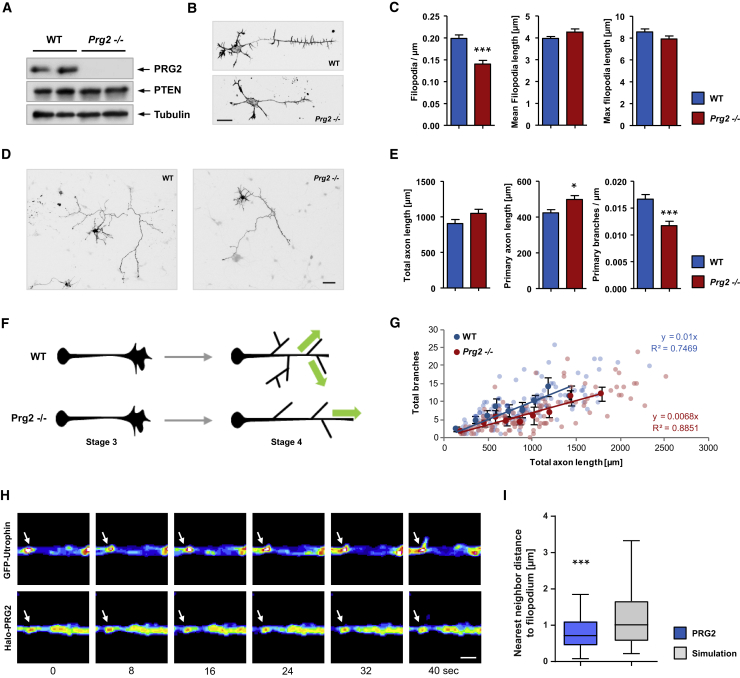
(A) Validation of Prg2 knockout (Prg2−/−) mouse by WB using P8 brain lysates.
(B) Hippocampal neurons from WT or Prg2−/− mice imaged at 1.5 DIV. Scale bar: 20 μm.
(C) Bars show the mean number of filopodia per axon length, the mean length of filopodia, and the mean length of the longest filopodium/neuron ± SEM of four independent experiments, n ≥ 100. ∗∗∗p < 0.001 (t test).
(D) Hippocampal neurons from WT or Prg2−/− mice imaged at 5 DIV. Scale bar: 50 μm.
(E) Bar graphs show mean ± SEM of 3 independent experiments, n ≥ 100. ∗p < 0.05, ∗∗∗p < 0.001 (t test).
(F) Schematic representation of Prg2−/− neuronal growth at two developmental stages.
(G) Correlation of branches and total axon length. Transparent dots represent individual neurons of either WT (blue) or Prg2−/− (red); darker dots represent means for equal clusters of neurons to reduce the complexity of the dataset.
(H) Representative filopodium emerging from a local PRG2 spot. Panels show F-actin probe GFP-Utrophin and Halo-tagged PRG2 over 40 s in 16-color Look-Up Table (LUT; FIJI), scale bar: 2 μm.
(I) Boxplots of nearest-neighbor analyses show smaller distances of emerging filopodia to PRG2 clusters when compared with randomly distributed clusters. Whiskers show the 5%–95% interval, ∗∗∗p < 0.001 (Wilcoxon signed rank test), n = 73 from four independent experiments.
See also Figures S5 and S6.