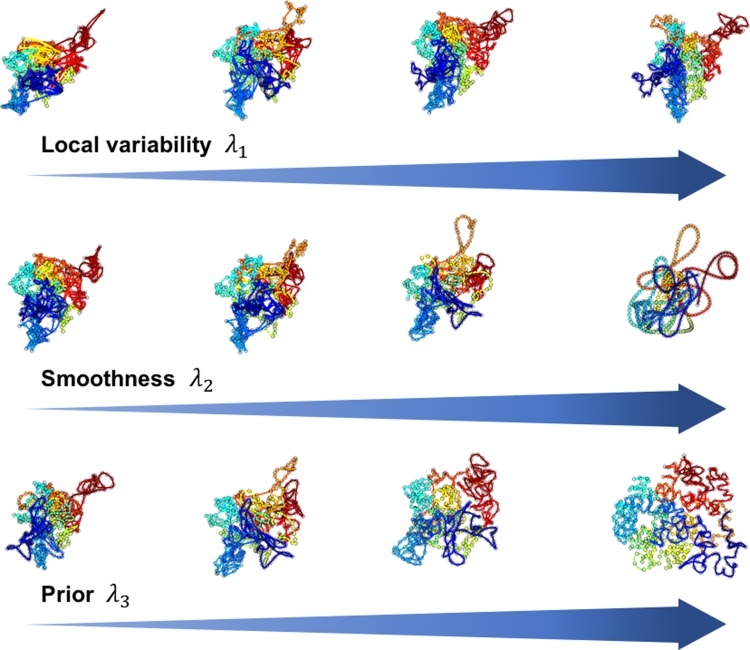
FIG. 5.
Effect of model parameters on 3D reconstruction quality. We compute the 3D reconstruction as a function of parameter values using the Hi-C data matrix associated with chromosome 19 in cell 1 of the mESC data set. The top row of structures from left to right shows the effect of an increased weight λ1 on the parameterization penalty h1. We vary λ1 = 0.01, 0.1, 1, 10 and fix λ2 = λ3 = 0 to obtain four solution curves with exponentially increasing penalty weight. The center row of structures from left to right shows the effect of an increasing weight λ2 on the smoothing penalty h2. Here we vary λ2 = 0.01, 0.1, 1, 10 and fix λ1 = 0.5 and λ3 = 0 to obtain these four solution curves. Finally, to show the effect of incorporating the bulk data—the mESC chromosome 19 population matrix—in the analysis, we vary λ3 = 0.01, 0.1, 1, 10 and fix λ1 = 0.5, λ2 = 1 to obtain the four structures on the bottom row. We computed all structures using the multiscale approach with n = 73, 146, 292, 584. 3D, three-dimensional; mESC, mouse embryonic stem cell.