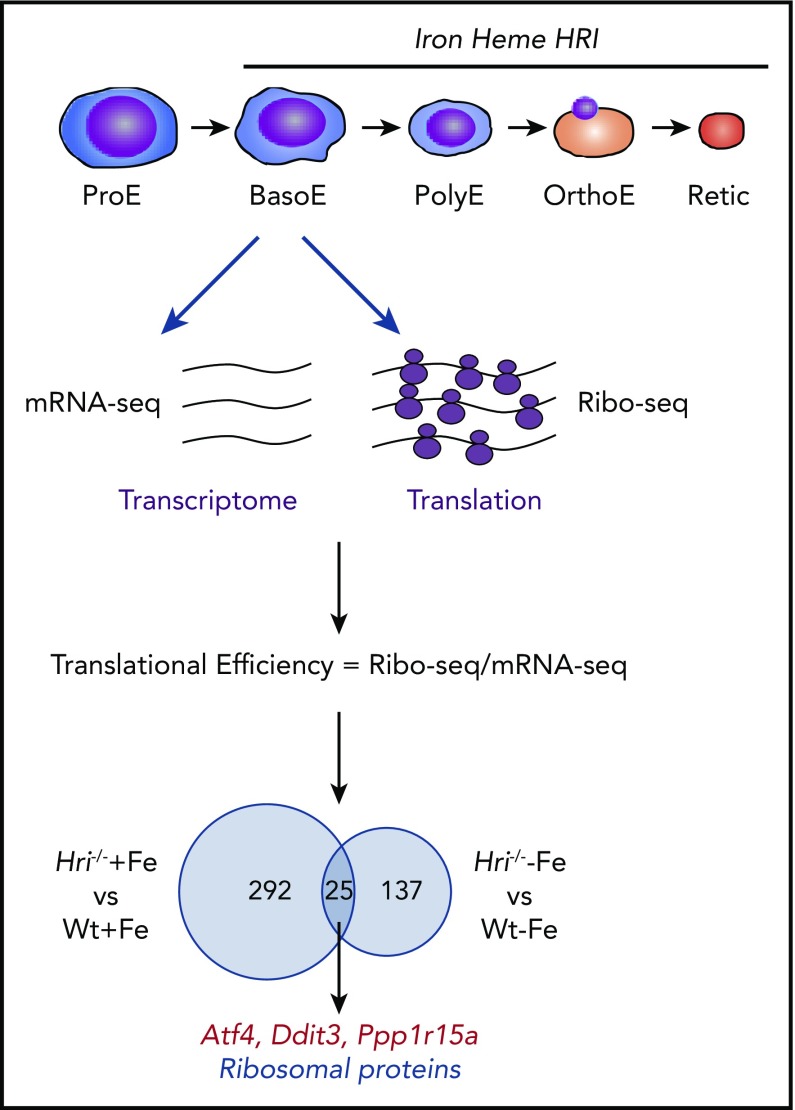
Figure 3.
Genome-wide in vivo translation assessment by ribosome profiling in iron and HRI deficiencies. Iron, heme, and HRI are critical for terminal erythropoiesis from Baso erythroblasts (BasoE) to reticulocytes (Retic). BasoE were sorted by flow cytometry using CD71 and Ter119 surface markers from embryonic day 14.5 FLs of Wt and Hri−/− embryos under +Fe and −Fe conditions. Actively translated mRNAs are occupied by ribosomes, which protect fragments of mRNAs from RNase digestion. cDNA libraries from ribosome-protected fragments and polyA+ mRNAs were subjected to DNA sequencing, followed by mapping to mouse genome mm10.46 Translational efficiency (TE) of each mRNA was calculated as the ratio of Ribo-seq reads/mRNA-seq reads. Venn diagrams of numbers of differential translated mRNAs between Wt and Hri−/− BasoEs under +Fe and −Fe are shown. Under both +Fe and −Fe conditions, TE of Atf4 mRNA is most highly enhanced in Wt BasoEs compared with Hri−/− cells, whereas translation of ribosomal protein mRNAs is most highly enhanced in Hri−/− BasoEs compared with Wt cells. Adapted from Zhang et al with permission.46