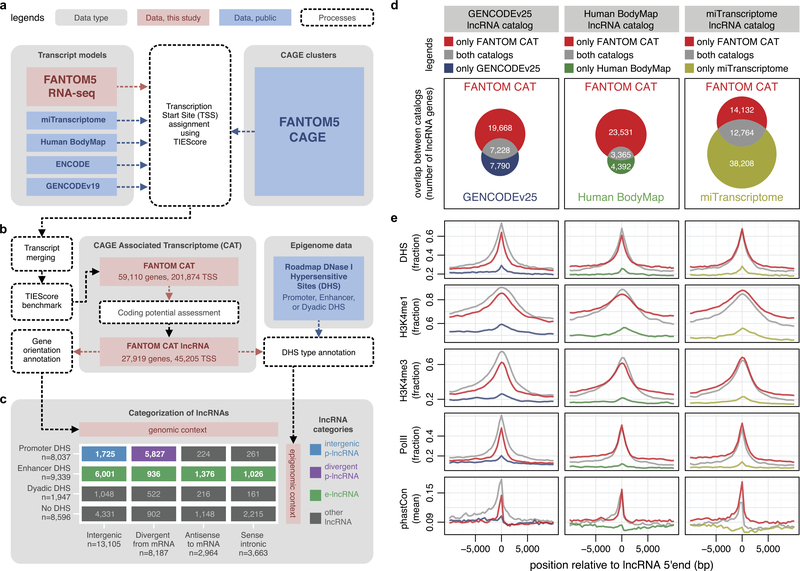
Extended Data Figure 1 |. Building a 5′ complete lncRNA catalogue.
a, Integration of CAGE and transcript models. CAGE clusters were used to integrate transcript models from various sources and their 5′ completeness was assessed on the basis of TIEScore. b, Identification of lncRNAs. TIEScore identified 59,110 genes and coding potential assessment further identified 27,919 lncRNAs in FANTOM CAT at the robust TIEScore cutoff. c, Categorization of lncRNAs. LncRNAs were annotated according to their gene orientation (that is, genomic context) and DHS type23 (that is, epigenomic context) and then categorized into divergent p-lncRNAs (purple), intergenic p-lncRNAs (blue), e-lncRNAs (green) and other lncRNAs (grey). d, Overlaps between FANTOM CAT and other lncRNA catalogues. e, LncRNA gene models outside FANTOM CAT are 5′ incomplete. LncRNAs found commonly in both catalogues (grey), or only in FANTOM CAT (red), show stronger evidence of transcription initiation (DHS, H3K4me1, H3K4me3 and PolII ChIP-seq23) and conservation (phastCons38) than those found only in other lncRNA catalogues (blue, green or yellow).