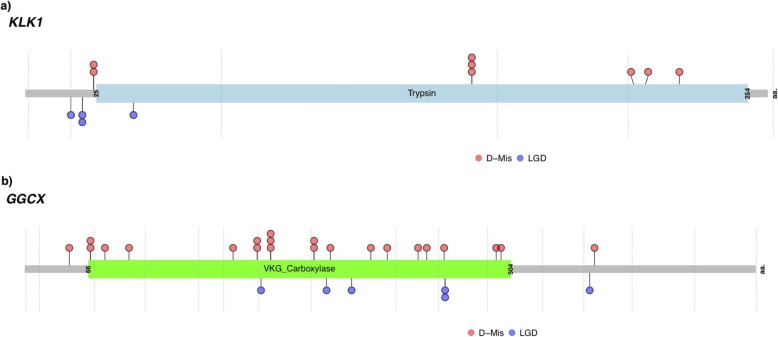
Fig. 5.
Locations of rare, predicted deleterious variants in KLK1 (a) and GGCX (b) across the PAH Biobank cohort (n = 2572 cases). Locations are provided within the two-dimensional protein structures. The numbers of variants at each amino acid position are indicated along the y-axes. The vertical gray lines indicate exon borders. D-MIS, predicted damaging missense; LGD, likely gene disrupting (stop-gain, frameshift, splicing)