Abstract
Small cell carcinoma of the ovary, hypercalcemic type (SCCOHT) is a rare but aggressive and untreatable malignancy affecting young women. We and others recently discovered that SMARCA4, a gene encoding the ATPase of the SWI/SNF chromatin-remodeling complex, is the only gene recurrently mutated in the majority of SCCOHT. The low somatic complexity of SCCOHT genomes and the prominent role of the SWI/SNF chromatin-remodeling complex in transcriptional control of genes suggest that SCCOHT cells may rely on epigenetic rewiring for oncogenic transformation. Herein, we report that approximately 80% (19/24) of SCCOHT tumor samples have strong expression of the histone methyltransferase EZH2 by immunohistochemistry with the rest expressing variable amounts of EZH2. Re-expression of SMARCA4 suppressed the expression of EZH2 in SCCOHT cells. In comparison to other ovarian cell lines, SCCOHT cells displayed hypersensitivity to EZH2 shRNAs and two selective EZH2 inhibitors, GSK126 and EPZ-6438. EZH2 inhibitors induced cell cycle arrest, apoptosis, and cell differentiation in SCCOHT cells, along with the induction of genes involved in cell cycle regulation, apoptosis and neuron-like differentiation. EZH2 inhibitors suppressed tumor growth and improved the survival of mice bearing SCCOHT xenografts. Therefore, our data suggest that loss of SMARCA4 creates a dependency on the catalytic activity of EZH2 in SCCOHT cells and that pharmacological inhibition of EZH2 is a promising therapeutic strategy for treating this disease.
Keywords: SCCOHT, SWI/SNF, chromatin remodeling complex, differentiation, EZH2, SMARCA4, ovarian cancer
Introduction
Small cell carcinoma of the ovary, hypercalcemic type (SCCOHT), a rare but highly aggressive ovarian malignancy with unknown cellular origin, occurs both sporadically and in families [1,2]. Unlike most common ovarian cancers, SCCOHT primarily affects women in their teens and twenties [3–5]. Surgical debulking followed by adjuvant chemotherapy is the mainstay of therapy for SCCOHT. However, recurrence is generally rapid, and the prognosis is dismal – about two-thirds of patients with advanced stage disease die within two years of diagnosis [4,6]. Therefore, more effective treatment options are urgently needed. Despite that Otte et al. have recently discovered c-Met inhibitors as potential targeted therapy agents for a subset of SCCOHT [7], the biology-driven therapeutic options for SCCOHT remain to be explored.
Several research teams, including ourselves, have independently identified inactivating, often homozygous or bi-allelic, mutations of the SMARCA4 gene in over 90% of SCCOHT cases, which leads to loss of SMARCA4 protein in the majority of SCCOHT tumors and cell lines [8–11]. Unlike common malignancies, no recurrent somatic, non-silent mutations besides those in SMARCA4 have been detected by paired exome or whole-genome sequencing analysis in SCCOHT [8,10–12]. Therefore, the inactivating mutations in SMARCA4 appear to be the primary driver in SCCOHT tumorigenesis and may help inform novel treatment strategies for SCCOHT.
SMARCA4 is one of the two mutually exclusive ATPases of the SWI/SNF multi-subunit chromatin-remodeling complex, which uses ATP hydrolysis to destabilize histone-DNA interactions and mobilize nucleosomes. The SWI/SNF complex localizes near transcriptional regulatory elements and regions critical for chromosome organization to regulate the expression of many genes involved in cell cycle control, differentiation and chromosome organization [13,14]. Several subunits of the SWI/SNF complex, such as SMARCA4, SMARCB1, ARID1A, PBRM1, are frequently mutated and inactivated in a variety of cancers [14–16]. This highlights the broader potential utility of effective targeted therapies for patients with a defective SWI/SNF complex. Recently, several studies reported that SMARCA4-deficient lung cancer cell lines relied on the activities of SMARCA2, the mutually exclusive ATPase, for proliferation [17,18], raising the possibility of selectively targeting SMARCA2 as therapeutic approaches for these patients. However, all SMARCA4-negative SCCOHT tumors and tumor-derived cell lines also lack the expression of SMARCA2 without apparent mutations in the SMARCA2 gene [19], indicating the need for developing different biologically informed treatment approaches for SCCOHT.
The interplay between the SWI/SNF complex and the Polycomb repressive complex 2 (PRC2) was originally demonstrated through genetic studies in Drosophila [20]. Mouse studies revealed that tumorigenesis driven by SMARCB1 loss was ablated by the simultaneous loss of EZH2, the catalytic subunit of PRC2 that trimethylates lysine 27 of histone H3 (H3K27me3) to promote transcriptional silencing [21]. Therefore, EZH2 has emerged as a putative therapeutic target for SMARCB1-deficient malignant rhabdoid tumors (MRTs), ARID1A-deficient ovarian clear cell carcinomas, SMARCA4-deficient lung cancers and PBRM1-deficient renal cancers, although the non-catalytic activity of EZH2 was likely responsible for the therapeutic potential in some cases [21–23]. Therefore, we set out to address whether targeting EZH2 is a feasible strategy for treating SMARCA4-deficient SCCOHT. We discovered that EZH2 is abundantly expressed in SCCOHT and its inhibition robustly suppressed SCCOHT cell growth, induced apoptosis and neuron-like differentiation, and delayed tumor growth in mouse xenograft models of SCCOHT.
Materials and methods
Cell culture and chemicals
Cells were cultured in either DMEM/F-12 (BIN67, SCCOHT-1 and COV434) or RPMI (all other lines) supplemented with 10% FBS and maintained at 37 °C in a humidified 5% CO2-containing incubator. All cell lines have been certified by STR analysis, tested regularly for Mycoplasma and used for the study within six months of thawing. EPZ-6438 and GSK126 were purchased from Selleckchem (in vitro studies) and Active Biochemku (in vivo studies).
Proteomics
Cells were lysed in 100mM HEPES buffer (pH 8.5) containing 1% SDS and 1x protease inhibitor cocktail (Roche). After chromatin degradation by benzonase, reduction and alkylation of disulfide bonds by dithiothreitol and iodoacetamide, samples were cleaned up and prepared for trypsin digestion using the SP3-CTP method [24]. In brief, proteins were digested for 14 h at 37 °C followed by removal of SP3 beads. Tryptic peptides from each sample were individually labeled with TMT 10-plex labels, pooled and fractionated into 12 fractions by high pH RP-HPLC, desalted, orthogonally separated and analyzed using and Easy-nLC 1000 coupled to a Thermo Scientific Orbitrap Fusion mass spectrometer operating in MS3 mode. Raw MS data were processed and peptide sequences were elucidated using Sequest HT in Proteome Discoverer software (v2.1.0.62), searching against the UniProt Human Proteome database.
Mouse xenografts
Animal handling, care, and treatment procedures were performed according to guidelines approved by the Animal Care Committee of the University of British Columbia (A14–0290). In brief, BIN67 (1×107 cells/mouse) or SCCOHT-1 cells (4×106 cells/mouse) were injected with a 1:1 mix of Matrigel (Corning) in a final volume of 200 μl subcutaneously into the back of NRG (NOD.Rag1KO.IL2RγcKO) mice. Mice were randomized to treatment arms once the average tumor volume reached 100mm3. EPZ-6438 and GSK126 were formulated in 0.5% NaCMC (Sigma) containing 0.1% Tween-80 and 20% Captisol (CyDex Pharmaceutical, pH 4.5), respectively. In the BIN67 xenograft model, EPZ-6438 was administrated orally twice daily (BID, 0800/1600h) for eight d at 100 or 200 mg/kg, halted for six d, and then resumed at once daily (QD) dosing for additional three weeks or until humane endpoints were reached (i.e. tumor volume ~800 mm3), while GSK126 was dosed intraperitoneally at 150 mg/kg once daily (M-Sat) for three weeks. In SCCOHT-1 xenograft model, EPZ-6438 was orally administrated at 200 mg/kg once daily for three weeks or until reaching humane endpoints. Tumor volume and mouse weight were measured thrice weekly. Tumor volume was calculated as length*(width)2*0.52.
Statistical analysis
Student’s t-test was used to evaluate the significant difference between two groups in all experiments except proteomics. The Peptide Expression Change Averaging (PECA) analysis [25] was performed for comparing the proteomic profiles of BIN67 cells treated with or without EPZ-6438 to generate signal log ratio values (log fold-change), p-values and false discovery rate (fdr) adjusted p-values (p.fdr) using peptide level signal values. The PECA analysis [25] was performed for comparing the proteomic profiles of BIN67 cells treated with or without EPZ-6438. Survival curves and IC50 of drug treatment were determined by PRISM software. A p-value or p.fdr-value (for proteomics data) < 0.05 was considered significant.
Additional Material and Methods are available in the Supplementary Information online.
Results
EZH2 is expressed abundantly in SCCOHT
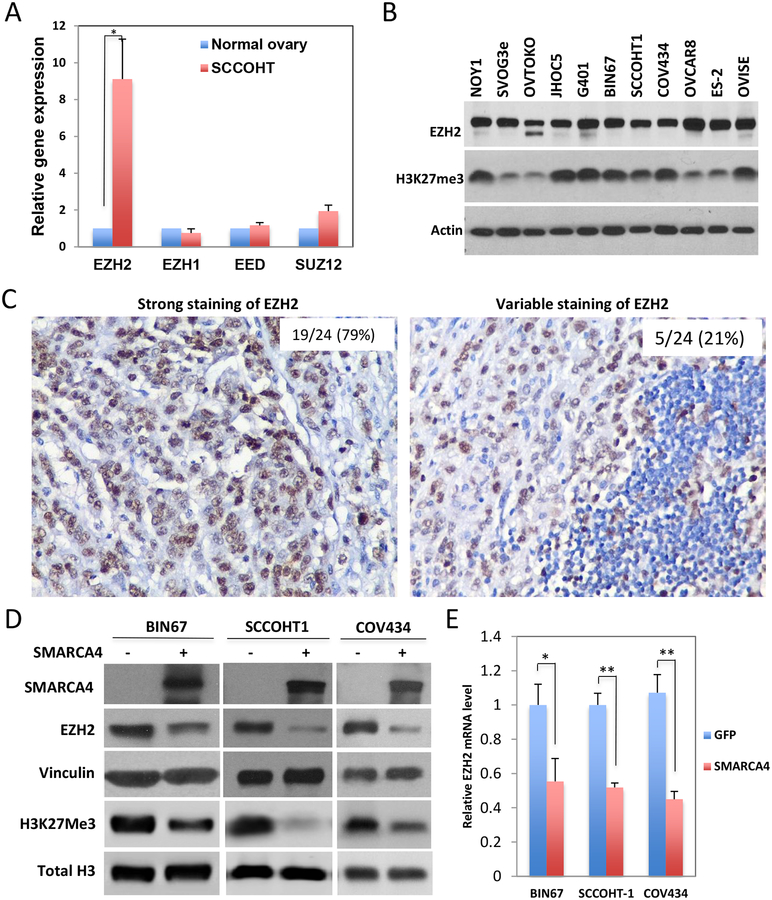
The poorly differentiated state of SCCOHT and the critical role of EZH2 in stem cell maintenance [26], together with the antagonism between the SWI/SNF complex and EZH2 [23], imply that EZH2 may play a role in SCCOHT tumorigenesis. Gene expression analysis of microarray data of four primary SCCOHT samples and two normal ovaries [19] showed that the expression of EZH2, but not other polycomb group (PcG) proteins, was significantly elevated in SCCOHT tumors compared to normal ovaries (Figure 1A). Western blotting analysis demonstrated that two SCCOHT cell lines, BIN67 and SCCOHT-1, expressed EZH2 protein at a level comparable to or higher than that several other ovarian cancer cell lines and the MRT cell line G401 (Figure 1B). COV434, a cell line originally designated as a juvenile granulosa cell tumor cell line [27] but recently redefined as a SCCOHT cell line with SMARCA4/SMARCA2 dual deficiency [28], also abundantly expressed EZH2 protein (Figure 1B). The expression levels of H3K27me3 were remarkably high in all three SCCOHT lines and G401, while only 3 out of 7 other ovarian cancer lines expressed H3K27me3 at similar abundance. Immunohistochemistry (IHC) analysis of a TMA of 24 primary SCCOHT tumors [19] revealed that 79% (19/24) displayed strong diffuse EZH2 staining with variable staining in the remainder (Figure 1C). Furthermore, EZH2 was substantially downregulated by SMARCA4 re-expression at both protein and RNA levels alongside a reduction in H3K27me3 levels in all three SCCOHT cell lines (Figure 1D and Figure 1E). These data suggest that loss of SMARCA4 correlates with upregulation of EZH2 and H3K27me3 levels in SCCOHT.
Figure 1. EZH2 is expressed abundantly in SCCOHT.
(A) The expression of PRC2 complex proteins in three SCCOHT versus two normal ovaries were extracted out from our previous Agilent microarray analyses (GE access No.: GSE49887 and GSE66434). (B) The expression levels of EZH2 and H3K27Me3 in multiple cell lines were analyzed by Western blotting analysis. (C) The expression of EZH2 protein in primary SCCOHT samples was determined using immunohistochemistry on TMA. Representative images of strong or variable staining are shown. (D, E) SCCOHT cell lines were infected with lentivirus expressing either GFP or SMARCA4. Cells were harvested 72 h post infection for determining the effect of SMARCA4 re-expression on EZH2 and histone H3 K27me3 by Western blotting (D) or the mRNA level of EZH2 by RT-qPCR (E). * P<0.05, ** P<0.001.
SCCOHT cells are sensitive to EZH2 inhibition
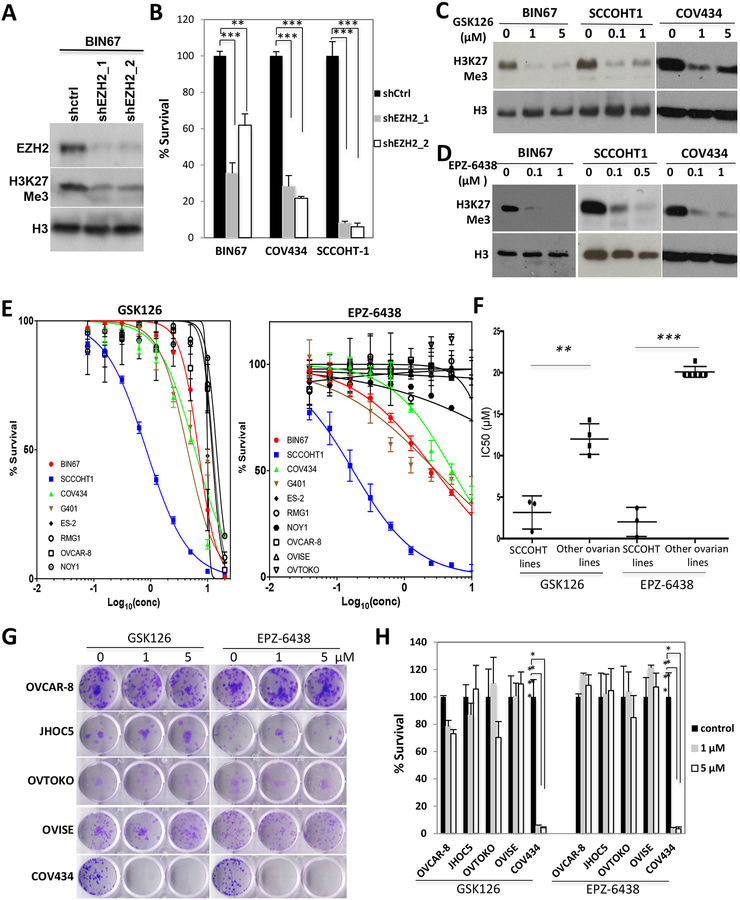
To determine whether SCCOHT relies on EZH2 for proliferation, we ablated the expression of EZH2 with two specific shRNAs. Depletion of EZH2 significantly reduced H3K27me3 levels (Figure 2A) and inhibited the growth of not only three SCCOHT cell lines (Figure 2B) but also several ovarian epithelial cancer cell lines (OVCAR-8, OVISE, RMG1 and JHOC5) with a mild effect in ES-2 cells (supplementary information, Figure S1), suggesting that EZH2 is required for the growth of many ovarian cancer cell lines.
Figure 2. SCCOHT cells are sensitive to EZH2 depletion and suppression.
(A, B) Cells were infected with lentivirus expressing control scramble shRNA or EZH2 shRNA followed by puromycin selection for 48 h. Cells were then reseeded in 24-well plates for 6 d before being fixed and quantitated by crystal violet staining assay. (C, D) Cells were treated as indicated for 3 d for Western blot analysis of histone H3K27me3 level. (E) Cells were seeded in 96-well plates, treated with EZH2 inhibitors GSK126 or EPZ-6438 at indicated doses and incubated for 6 d and measured for cell survival by crystal violet assay. (F) The IC50s of cell lines to EZH2 inhibitors in (E) were compared between SCCOHT lines and other ovarian lines. Note: 20 μM was assigned to the cell lines that were not responsive to EZH2 inhibitors at 10 μM. (G, H) Cells were seeded in 12-well plates, treated with EZH2 inhibitors GSK126 or EPZ-6438 at indicated doses for 14 d and measured for clonogenic abilities by crystal violet assay. ** P<0.01, *** P<0.001
Next, we determined whether SCCOHT cells are responsive to the catalytic inhibition of EZH2 with two pharmaceutical inhibitors, GSK126 and EPZ-6438, which potently suppressed histone H3K27me3 levels (supplementary information, Figure S2, Figure 2C, D). In a six-day drug treatment assay, all SCCOHT lines were significantly more sensitive to either GSK126 or EPZ-6438 than other ovarian cancer lines tested (Figure 2E, F). Among SCCOHT lines, SCCOHT-1 cells were about 5 or 10-fold more sensitive than BIN67, COV434 and the MRT cell line G401 to the treatment of GSK126 or EPZ-6438, respectively (Figure 2E). The drug response difference between SCCOHT lines and other ovarian cancer lines became more drastic in 9-day survival assays (supplementary information, Figure S3). Clonogenic assays confirmed that only SCCOHT cells were responsive to either 1 or 5 μM of GSK126 or EPZ-6438 treatment (Figure 2G) and 0.1 μM of EPZ-6438 (supplementary information, Figure S4) for 2 weeks. However, neither SMARCA4- nor SMARCA4/SMARCA2-dual-deficient lung tumor cells were as sensitive as SCCOHT cells in 6-day survival assays (supplementary information, Figure S5).
EPZ-6438 suppresses tumor growth in mouse SCCOHT xenograft models
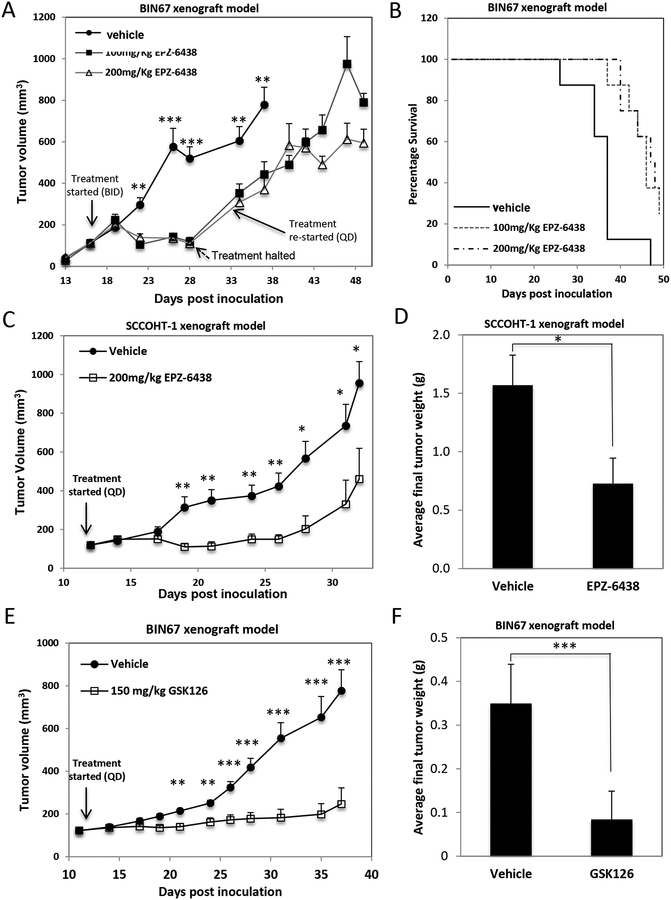
Next, we employed mouse subcutaneous xenografts of SCCOHT cells to evaluate in vivo efficacy of EZH2 inhibitors. Twice daily treatment with EPZ-6438 (100 or 200 mg/kg, n=10) [29–31] for 8 days significantly lowered the H3K27me3 level of BIN67 xenograft tumors (supplementary information, Figure 6A) and robustly suppressed tumor growth compared to those treated with vehicle (supplementary information, Figure S6B, day 28, P <0.01, P <0.01, respectively) along with significant body weight loss (supplementary information, Figure S6C). Dosing was halted for 6 days to allow recovery from dehydration and resumed with daily single dose schedules until mice reached humane endpoints, which was well tolerated but failed to delay tumor growth strongly (supplementary information, Figure S6B, days 34–48). The median survival of mice to humane endpoint was increased from 37 days (vehicle) to 46 days (100mg/kg) and 48 days (200 mg/kg) mainly due to the benefit from twice daily dosing (supplementary information, Figure S6D, P = 0.0036 and 0.0017, respectively).
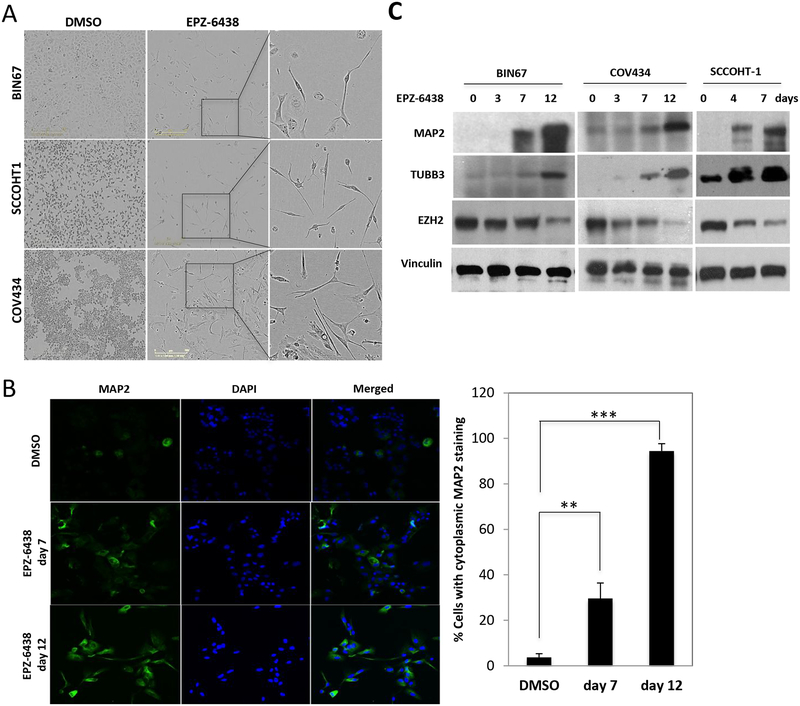
Figure 6. EPZ-6438 induces neuronal differentiation in SCCOHT cells.
(A) Morphology of SCCOHT cells after prolonged exposure to EPZ-6438. Cells were treated with EPZ-6438 at either 1 μM (BIN67/COV434) or 0.25 μM (SCCOHT-1) for 12 days and characterized by phase contrast microscopy. (B) BIN67 cells were fixed and immunostained for MAP2, a selective neuronal marker. Percentage of cells with cytoplasmic staining of MAP2 was quantitated. (C) Western blotting for neuronal proteins from SCCOHT cells treated with EPZ-6438 at either 1 μM (BIN67/COV434) or 0.25 μM (SCCOHT-1) for the days shown in the panel. Vinculin served as a loading control. ** P<0.01, *** P<0.001
Next, we tested the efficacy of EPZ-6438 in the SCCOHT-1 xenograft model with a 200 mg/kg EPZ-6438 once daily schedule (n=6) due to the high potency of EZH2 inhibitors in SCCOHT-1 cells in vitro. Treatment of EPZ-6438 for 3 weeks mildly affected the mouse body weight (supplementary information, Figure S7A) and effectively suppressed tumor growth with the doubling time (tumor volume from 150 mm3 to 300 mm3) increased from 5 days (vehicle) to 17 days (EPZ-6438) (Figure 3A). The average tumor weight was also reduced by ~50% in the EPZ-6438-treated group compared to the vehicle group at the end of the treatment (Figure 3B, p < 0.05). These data suggest that once daily treatment of EPZ-6438 at 200 mg/kg significantly slowed down the growth of SCCOHT-1 xenografts.
Figure 3. In vivo efficacy of EPZ-6438 in BIN67 mouse xenograft model.
(A-B) The efficacy of EPZ-6438 was evaluated in SCCOHT-1-derived mouse subcutaneous xenograft model. Tumor volume of either vehicle or 200 mg/kg EPZ-6438-treated group was plotted against the days post cell inoculation (A). Final tumor weight was determined and compared between two treated groups (B). (C, D) The efficacy of GSK126 was evaluated in BIN67 mouse subcutaneous xenograft model. Tumor volume of either vehicle or 150 mg/kg GSK126-treated group was plotted against the days post cell inoculation (C) with final tumor weights being compared between two treated groups (D). * P<0.05, ** P<0.01, *** P<0.001
Last, we further examined the efficacy of GSK126 in BIN67 xenograft model. Once daily dosing of GSK126 at 150 mg/kg (n=8) for three weeks had no effect on mouse body weight gain (supplementary information, Figure S7B) but robustly suppressed the tumor growth (Figure 3C). The average tumor weight dropped by ~75% in the GSK126-treated group compared to the vehicle group at the end of the treatment (Figure 3D, p < 0.001).
Effects of EPZ-6438 on the proteome of BIN67 cells
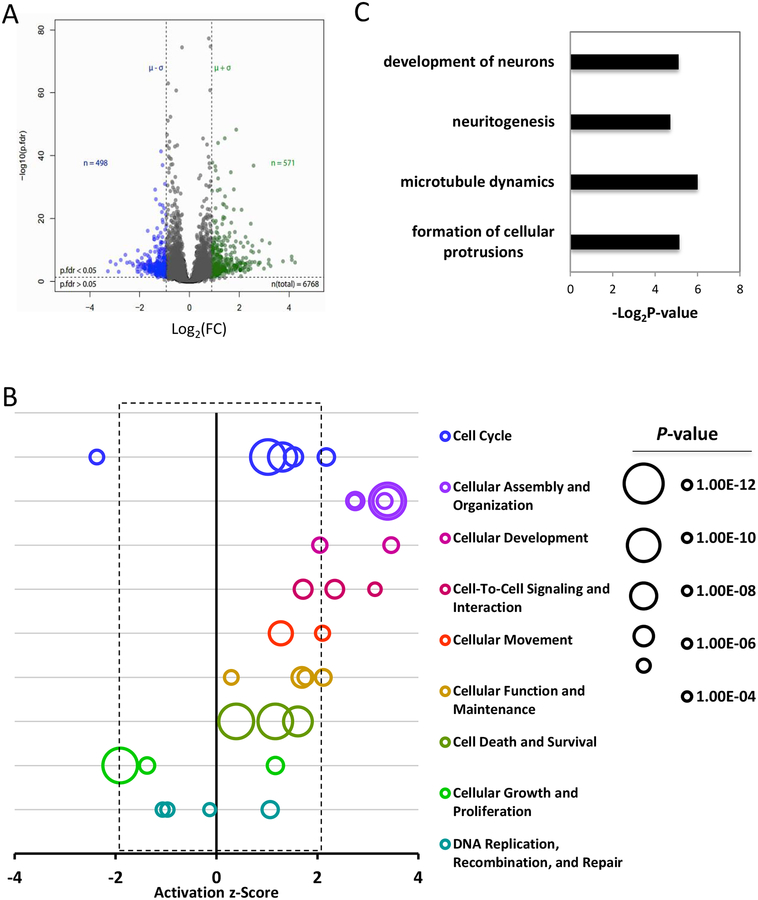
To identify the molecular mechanisms driving growth inhibition caused by EZH2 inhibitors in SCCOHT cells, we determined the proteomic profiles of BIN67 cells treated with EPZ-6438 or vehicle using mass spectrometry. Unsupervised hierarchical clustering resulted in clustering based on the treatment conditions (supplementary information, Figure S8) with approximately 8.4% (571/6768) and 7.4% (498/6768) being significantly upregulated or downregulated, respectively, in BIN67 cells treated with 1 μM EPZ-6438 for 7 days versus those treated with DMSO (Figure 4A and supplementary information, Tables S1 and S2, pfdr<0.05 and log2FC>mean+SD or <mean-SD). Ingenuity pathway analysis (IPA) of the significantly altered proteins by EPZ-6438 revealed a significant enrichment of proteins involved in biological functions, such as “Cell cycle”, “Cellular Assembly and Organization” and “Cellular development” (Figure 4B, supplementary information, Table S3). Particularly, both “development of neurons” and several processes related to neuronal development, such as “formation of cellular protrusions”, “microtubule dynamics” and “neuritogenesis”, were predicted to increase significantly in EPZ-6438-treated BIN67 cells (Figure 4C, supplementary information, Table S3, z>2). Clustering of the enriched proteins confirmed that many of the identified proteins involved in microtubule dynamics, formation of cellular protrusions and organization of cytoskeleton or cytoplasm were also involved in development of neurons (supplementary information, Figure S9), supporting their engagement in neuron development. These data suggest that EPZ-6438 not only altered the cell cycle of SCCOHT cells, but also triggered significant changes in cell organization and assembly, leading to neuron-like differentiation.
Figure 4. The effect of EPZ-6438 on the proteome of BIN67 cells.
(A) Volcano plot of the proteome of BIN67 cells exposed to EPZ-6438 or vehicle. Cells were treated with either DMSO or 1 μM EPZ-6438 for 7 d and then processed for proteomic profiling. Peptide data were subjected to PECA analysis for identification of significantly altered proteins (p.fdr<0.05 and Log2FC>mean+SD or <mean-SD). (B) IPA analysis of significantly altered proteins caused by EPZ-6438 identified top affected biological functions by EPZ-6438 treatment. Any biological function with an activation z-score greater than 2 or less than −2 was predicted to be significantly increased or decreased by IPA analysis. (C) IPA analysis identified significantly increased cellular activities related to neuronal development. (D) Clustering analysis of proteins in significantly altered biological functions.
EPZ-6438 induces SCCOHT cell cycle arrest and apoptosis
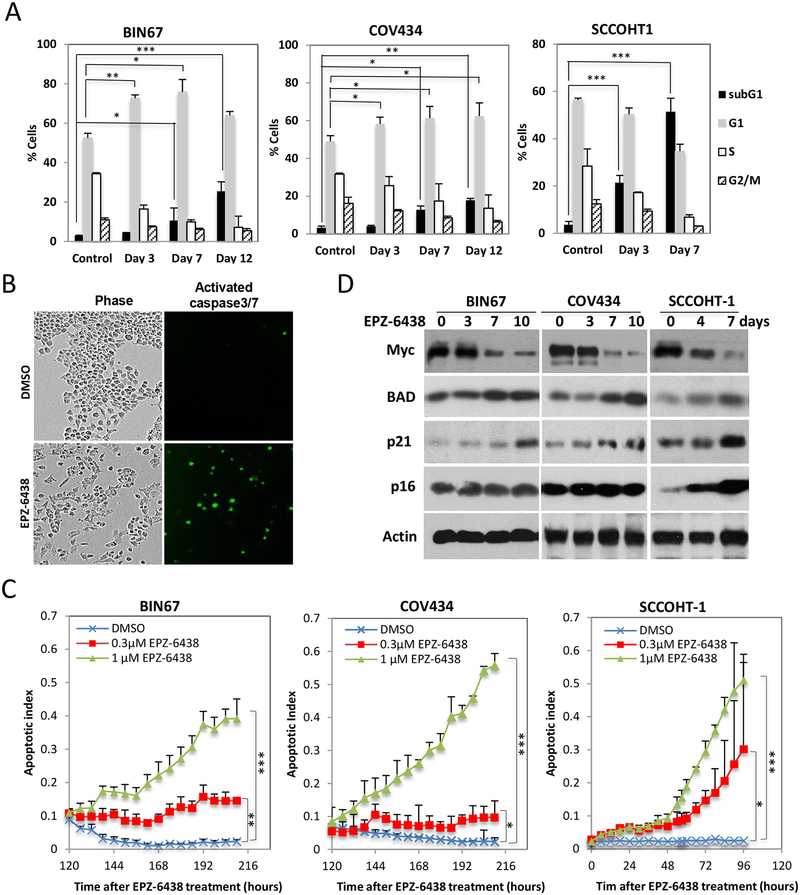
Next, we used flow cytometry to validate the effect of EPZ-6438 on cell cycle distribution of SCCOHT cells. FACS analysis demonstrated that 1 μM EPZ-6438 treatment induced cell cycle arrest at G1 in a time-dependent manner in BIN67 and COV434 cells (Figure 5A), and also triggered cell death (sub-G1 population) beginning at 7 days after drug exposure in both BIN67 and COV434 cell lines, consistent with a mild induction of apoptosis predicted by proteomic data (supplementary information, Table S4). In contrast, 1 μM EPZ-6438 induced cell death more rapidly (3 days) and potently in SCCOHT-1 cells (Figure 5A), consistent with the lower IC50 in this cell line than others (Figure 2E). To precisely monitor the induction of apoptosis, we measured the activation of caspase-3/7 through fluorescent microscopy coupled with live cell imaging (Figure 5B). This revealed that EPZ-6438 rapidly induced cell apoptosis in SCCOHT-1 cells after 48 hours of EPZ-6438 treatment at either 0.3 or 1 μM, whereas apoptosis in BIN67 and COV434 cells increased gradually beginning at about 120 hours of exposure to 1 μM EPZ-6438 (Figure 5C). Next, we employed western blotting to determine the expression of some cell cycle and apoptosis regulating genes that were significantly altered by EPZ-6438 in BIN67 cells by proteomic profiling. We confirmed that the expression of CDKN1A (p21) and BAD (Log2FC =0.7 and 1.4, respectively, supplementary information, Table S1) was gradually induced by EPZ-6438, while that of Myc (Log2 FC = −0.9, supplementary information, Table S2) was potently down-regulated by EPZ-6438 in all three SCCOHT cell lines (Figure 5D). In contrast, the expression of CDKN2A (p16) (Log2 FC = 0.2, supplementary information, Table S1), which is known to play an important role in EZH2 inhibitor-triggered growth arrest [32], was only significantly induced upon EPZ-6438 treatment in SCCOHT-1 cells (Figure 5D).
Figure 5. EPZ-6438 induces SCCOHT cell cycle arrest and apoptosis.
(A) FACS analysis of cell cycle profiles in SCCOHT cells after DMSO or 1 μM EPZ-6438 treatment for the times shown. (B, C) SCCOHT cell lines were treated with 1 μM EPZ-6438 and cultured in the presence of a cell-permeable fluorescent dye for monitoring activated caspase-3/7 activity with IncuCyte live cell imaging system (see Materials and Methods). Apoptotic index was calculated by dividing the overall fluorescent object counts to cell numbers under each condition and plotted over incubation time. (D) The effects of EPZ-6438 (BIN67/COV434: 1 μM; SCCOHT-1: 0.25 μM) on the expression of Myc, BAD, p16 and p21 proteins was determined by western blotting. * P<0.05, ** P<0.01, *** P<0.001
EPZ-6438 induces neuron-like differentiation in SCCOHT cells
In agreement with the prediction of induced neuron development in BIN67 cells from proteomic profiling, the surviving SCCOHT cells displayed a neuron-like morphology after prolonged exposure to EPZ-6438 (Figure 6A). In contrast, SCCOHT cells exposed to cytotoxic agents, such as cisplatin, etoposide, and paclitaxel, displayed no morphology change other than fragmentation during cell death (supplementary information, Figure S10). Immunofluorescence confirmed that the expression of MAP2, a specific marker of neuronal differentiation [33], was increased upon EPZ-6438 treatment in a time-dependent manner in BIN67 cells (Figure 6B), consistent with increased expression with treatment observed in the proteomic data (Log2 FC = 2.2, supplementary information, Table S2). Western blot analysis further confirmed that the expression levels of two neuronal markers (MAP2 and TUBB3 (βIII tubulin), Log2FC=1.2, supplementary information, Table S2) were induced by EPZ-6438 treatment (Figure 6C). In agreement with the induction of differentiation, the expression of EZH2 (Log2FC=−0.4, supplementary information, Table S1) also dropped significantly upon EPZ-6438 treatment in a time-dependent manner (Figure 6C). Therefore, our data show that unlike cytotoxic agents, EZH2 inhibitors can induce expression of markers of neuronal differentiation of SCCOHT cells.
Discussion
Transformation of normal cells requires acquisition of hallmark characteristics of cancer including survival and proliferation even in the presence of counteracting signals. These features usually include extensive rewiring of cellular signaling networks driven by mutations and deregulation of oncogenes and tumor suppressors [34], leading to a strict reliance on either an oncogenic driver event (oncogene addiction) or the activity of certain gene products that are not essential in normal cells (non-oncogene addiction or synthetic lethality) [35]. Our findings suggest that inactivation of SMARCA4 in SCCOHT’s unknown precursor cells may rewire their cellular signaling network to be dependent on the histone methyltransferase EZH2 in transcriptional repression. SMARCA4 and the SWI/SNF chromatin-remodeling complex can suppress EZH2 either directly [21] or through repression of E2F transcription factors [36] consistent with the re-expression of SMARCA4 lowering the expression of EZH2 in our SCCOHT cell lines (Figure 1D). However, expression levels of EZH2 do not predict cellular response to EZH2 inhibitors as observed in the other ovarian cancer cell lines (Figure 1B and Figure 2E). These results suggest that the PRC2 complex and the SWI/SNF complex antagonistically regulate cellular targets that determine the cellular response to EZH2 inhibitors in SCCOHT. Accordingly, recent studies demonstrated that SMARCA4 loss in mouse embryonic stem cells led to failed eviction of PRC1 and PRC2 complexes from their occupied chromatin sites [37,38], providing an insight into the antagonism between SWI/SNF complex and polycomb repressive complexes. Unlike SCCOHT lines, several ovarian cancer lines (OVCAR-8, RMG1, OVISE and JHOC5) were only sensitive to EZH2 depletion, but not catalytic inhibition of EZH2, suggesting a unique requirement for the catalytic activity of EZH2 during the oncogenic transformation of SCCOHT. Of note, EZH2 inhibitor treatment significantly reduced EZH2 levels in all cell lines in a time-dependent manner, likely reflecting their differentiation state as previously reported during neuronal stem cell differentiation [39] and in skin cancer stem cells treated with EZH2 inhibitors [40].
In addition to this work, several other studies have demonstrated the requirement for both the methyltransferase activity and non-catalytic function of EZH2 in cancers with a defective subunit of the SWI/SNF complex. However, the growth of two ARID1A-deficient ovarian clear cell carcinoma cell lines (OVISE and OVTOKO) and several lung cancer cell lines with SMARCA4 deficiency or SMARCA4/SMARCA2 dual deficiency was suppressed by depletion of EZH2, but not EZH2 catalytic inhibitors (Figure 2E; supplementary information, Figures S3 and S4). Although future genetic rescue experiments are required for definitively addressing the dependency of these cell lines on the catalytic or non-catalytic function of EZH2, it remains possible that the response to the EZH2 catalytic inhibitors in cells with a defective SWI/SNF complex may be modulated by additional genetic or epigenetic features that either prevent the disruption of PRC2 complex by EZH2 inhibitors, which appears to be essential for measurable responses to EZH2 inhibitors [23], or trigger the non-catalytic function of EZH2 [41]. Of note, most tumors with SMARCA4 or ARID1A loss usually display a higher mutation burden in comparison to the minimally disturbed genomes of either SCCOHT or MRT [8,10,11,42]. The lack of additional mutations in the latter tumors implicates a dependence upon epigenetic changes such as EZH2 overexpression. Thus, SMARCA4 inactivation in the absence of SMARCA2 may rewire the cellular signaling network to be dependent on EZH2-mediated oncogenesis while additional mutations in other cancers possibly negate this requirement.
Although the potency of EZH2 inhibitors has been demonstrated in several cancer types, the molecular mechanisms are still not well understood. EZH2 is the catalytic component of the PRC2 complex that mediates the transcriptional repression of targets by trimethylation of histone H3 at lysine residue 27 in their promoters. Therefore, the PRC2 complex could promote tumorigenesis by specifically repressing tumor-suppressor genes, including the major tumor suppressor locus CDKN2A, through recruitment of DNA methyltransferases. Accordingly, treatment with EZH2 inhibitors can rescue the expression of p16 in cancer cells, such as MRT cells and leukemia stem cells [29,43]. Unlike MRT cells, in only one of the three SCCOHT cell lines (SCCOHT-1) was p16 expression altered upon EPZ-6438 treatment. The other two cell lines expressed substantial amounts of p16, which were not further induced upon the EPZ-6438 treatment. Therefore, a different mechanism may underlie the efficacy of EPZ-6438 in the BIN67 and COV434 cell lines.
Our proteome analysis revealed that EPZ-6438 altered about 15% of the detected proteins in BIN67 cells with a significant enrichment of proteins involved in cell cycle control and the development of neurons. These results implicate these pathways as the major biological events behind the cellular response to EZH2 inhibitors in SCCOHT cells. Particularly, we discovered that the expression of several cell cycle control genes, such as MYC, was repressed by EPZ-6438. Although the PRC2 complex may upregulate the expression of these genes indirectly through suppressing the negative regulators of these genes, it has been suggested that the transcription of MYC can be activated directly by the PRC2 complex in glioblastoma cancer stem cells [44]. Given that Myc activation is a hallmark of tumor initiation and maintenance, suppression of Myc may have a significant contribution to the efficacy of EZH2 inhibitors in SCCOHT and other cancers, such as MRT and glioblastoma [44]. Furthermore, we also observed a time-dependent induction of BAD pro-apoptotic protein in three SCCOHT cells following EPZ-6438 treatment. By inactivating the function of the anti-apoptotic proteins Bcl-2 and Bcl-xl, BAD can promote apoptosis [45]. Our data, together with that of the previous study showing that BAD was upregulated upon EZH2 depletion by shRNA in lung cancer cell lines [46], suggest that it may play a key role in EZH2 inhibitor-triggered apoptosis. In addition to causing cell cycle arrest and apoptosis, prolonged exposure to EZH2 inhibitors also drove the differentiation of SCCOHT cells towards the neuronal lineage. Consistent with the neuron-like differentiation, EPZ-6438 caused a late induction of p21 (Figure 4F), a cyclin kinase inhibitor that may drive neural precursor cells into cycle exit and differentiation [47]. It will be interesting to determine whether depletion of p21 can prevent the neuron-like differentiation caused by EZH2 inhibitors.
A neuron-like morphology change has also been reported in G401 cells upon treatment with EPZ-6438 [29]. Both genomic analysis and mouse transgenic models have provided evidence that some MRT may arise from neural precursor cells [42,48]. Interestingly, some studies suggest that SCCOHT is the MRT of ovary. Therefore, our present finding, that EZH2 inhibition induced markers of neuronal differentiation of SCCOHT, suggest that SCCOHT may develop from multi-potent stem cells inside the ovary with the capability of undergoing neuronal differentiation. In support of this model, extensive IHC analysis of multiple sections of two SCCOHT cases previously revealed rare foci of immature teratoma [9], which mainly contain primitive neuroepithelium in their immature region [49]. Future studies are therefore needed to explore the possible cellular origin of SCCOHT from neural precursor cells.
SCCOHT is a rare but extremely aggressive disease for young women without effective treatments. Our present study demonstrated that the methyltransferase EZH2 can serve as a potential therapeutic target for this deadly disease. While our manuscript was in review, an independent manuscript confirming some of our findings was published [28]. Although both papers described the preclinical potency of EZH2 inhibition in SCCOHT, we substantiated this effect with two distinct EZH2 inhibitors for both in vitro and in vivo studies. In addition, our study analyzed EZH2 expression in primary tumors, determined the difference between EZH2 catalytic inhibition and genetic depletion in SCCOHT lines versus other ovarian cancer lines, provided proteomic profiling of cells exposed to EPZ-6438, and revealed the neuron-like differentiation of SCCOHT cells upon EZH2 inhibition. The interim result from a phase 1 clinical trial that two SCCOHT patients displayed a partial response or stable disease by RECIST criteria upon EPZ-6438 treatment [50] supports the therapeutic potential of EZH2 inhibitors and evokes the needs for identifying other targeted therapy strategies and determining the efficacy of combining EZH2 inhibitors with other putative therapies for SCCOHT treatment.
Supplementary Material
Table S1. Proteins identified by mass spectrometry in BIN67 cells treated with either DMSO or 1 uM EPZ-6438 for 7 d
Table S2. Differentially expressed proteins in BIN67 cells treated with EPZ-6438 versus DMSO
Table S3. Biological functions affected by EPZ-6438 treatment
Table S4. Proteins that contributed to the significantly altered biological functions by EPZ-6438 treatment
Figure S1. Effect of EZH2 depletion in ovarian cancer cell lines
Figure S2. Effect of EPZ-6438 on histone H3K27me3
Figure S3. Responses of ovarian cancer cell lines to EZH2 inhibitors in 9-day survival assays
Figure S4. EPZ-6438 suppressed clonogenic activity of COV434 cells
Figure S5. SMARCA4-deficient lung cancer cells and ovarian clear cell carcinoma cells are not sensitive to EPZ6438
Figure S6. Effect of EPZ-6438 in Bin67 xenograft models
Figure S7. Mouse body weight changes in BIN67 and SCCOHT-1 xenograft models for the EZH2 inhibitor efficacy studies
Figure S8. Unsupervised clustering analysis of proteins identified by mass spectrometry in BIN67 cells treated with DMSO or EPZ-6438 for 7 d (n=3)
Figure S9. Clustering analysis of proteins involved in each significantly altered biological function predicted by IPA analysis
Figure S10. Cytotoxic agents do not induce neuron-like morphologies in SCCOHT cells
Acknowledgements
We acknowledge the technical support of Sarah Maines-Bandiera, Winnie Yang, Galen Lambert, Christine Chow, Nicole Wretham, Dana Masin, Hong Yan, Jenna Rawji and Chris Ke-dong Wang. We thank Dr Barbara Vanderhyden and Dr Ralf Hass for providing BIN67 and SCCOHT-1 cells, respectively. This work was supported by research funds from the Canadian Cancer Society Research Institute (#703458, to D.G.H.) and the National Institute of Health (1R01CA195670–01, to D.G.H., J.T. and B.W.), the Terry Fox Research Institute Initiative New Frontiers Program in Cancer (D.G.H.), the British Columbia Cancer Foundation, the Marsha Rivkin Center for Ovarian Cancer Research, the Ovarian Cancer Alliance of Arizona, the Small Cell Ovarian Cancer Foundation, and philanthropic support to the TGen Foundation.
Footnotes
Conflict of interest: None of the authors report any conflicts of interest.
References:
- 1.Longy M, Toulouse C, Mage P, et al. Familial cluster of ovarian small cell carcinoma: a new mendelian entity? J Med Genet 1996; 33: 333–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martinez-Borges AR, Petty JK, Hurt G, et al. Familial small cell carcinoma of the ovary. Pediatr Blood Cancer 2009; 53: 1334–1336. [DOI] [PubMed] [Google Scholar]
- 3.Florell SR, Bruggers CS, Matlak M, et al. Ovarian small cell carcinoma of the hypercalcemic type in a 14 month old: the youngest reported case. Med Pediatr Oncol 1999; 32: 304–307. [DOI] [PubMed] [Google Scholar]
- 4.Estel R, Hackethal A, Kalder M, et al. Small cell carcinoma of the ovary of the hypercalcaemic type: an analysis of clinical and prognostic aspects of a rare disease on the basis of cases published in the literature. Arch Gynecol Obstet 2011; 284: 1277–1282. [DOI] [PubMed] [Google Scholar]
- 5.Young RH, Oliva E, Scully RE. Small cell carcinoma of the ovary, hypercalcemic type. A clinicopathological analysis of 150 cases. Am J Surg Pathol 1994; 18: 1102–1116. [DOI] [PubMed] [Google Scholar]
- 6.Witkowski L, Goudie C, Ramos P, et al. The influence of clinical and genetic factors on patient outcome in small cell carcinoma of the ovary, hypercalcemic type. Gynecol Oncol 2016; 141: 454–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Otte A, Rauprich F, von der Ohe J, et al. c-Met inhibitors attenuate tumor growth of small cell hypercalcemic ovarian carcinoma (SCCOHT) populations. Oncotarget 2015; 6: 31640–31658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ramos P, Karnezis AN, Craig DW, et al. Small cell carcinoma of the ovary, hypercalcemic type, displays frequent inactivating germline and somatic mutations in SMARCA4. Nat Genet 2014; 46: 427–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kupryjanczyk J, Dansonka-Mieszkowska A, Moes-Sosnowska J, et al. Ovarian small cell carcinoma of hypercalcemic type - evidence of germline origin and SMARCA4 gene inactivation. a pilot study. Pol J Pathol 2013; 64: 238–246. [DOI] [PubMed] [Google Scholar]
- 10.Witkowski L, Carrot-Zhang J, Albrecht S, et al. Germline and somatic SMARCA4 mutations characterize small cell carcinoma of the ovary, hypercalcemic type. Nat Genet 2014; 46: 438–443. [DOI] [PubMed] [Google Scholar]
- 11.Jelinic P, Mueller JJ, Olvera N, et al. Recurrent SMARCA4 mutations in small cell carcinoma of the ovary. Nat Genet 2014; 46: 424–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science 2013; 339: 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Euskirchen GM, Auerbach RK, Davidov E, et al. Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS genetics 2011; 7: e1002008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson BG, Roberts CW. SWI/SNF nucleosome remodellers and cancer. Nat Rev Cancer 2011; 11: 481–492. [DOI] [PubMed] [Google Scholar]
- 15.Shain AH, Pollack JR. The spectrum of SWI/SNF mutations, ubiquitous in human cancers. PLoS One 2013; 8: e55119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kadoch C, Hargreaves DC, Hodges C, et al. Proteomic and bioinformatic analysis of mammalian SWI/SNF complexes identifies extensive roles in human malignancy. Nat Genet 2013; 45: 592–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson BG, Helming KC, Wang X, et al. Residual complexes containing SMARCA2 (BRM) underlie the oncogenic drive of SMARCA4 (BRG1) mutation. Mol Cell Biol 2014; 34: 1136–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoffman GR, Rahal R, Buxton F, et al. Functional epigenetics approach identifies BRM/SMARCA2 as a critical synthetic lethal target in BRG1-deficient cancers. Proc Natl Acad Sci U S A 2014; 111: 3128–3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karnezis AN, Wang Y, Ramos P, et al. Dual loss of the SWI/SNF complex ATPases SMARCA4/BRG1 and SMARCA2/BRM is highly sensitive and specific for small cell carcinoma of the ovary, hypercalcaemic type. J Pathol 2016; 238: 389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kennison JA. The Polycomb and trithorax group proteins of Drosophila: trans-regulators of homeotic gene function. Annu Rev Genet 1995; 29: 289–303. [DOI] [PubMed] [Google Scholar]
- 21.Wilson BG, Wang X, Shen X, et al. Epigenetic antagonism between polycomb and SWI/SNF complexes during oncogenic transformation. Cancer Cell 2010; 18: 316–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bitler BG, Aird KM, Garipov A, et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med 2015; 21: 231–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim KH, Kim W, Howard TP, et al. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nat Med 2015; 21: 1491–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hughes CS, McConechy MK, Cochrane DR, et al. Quantitative Profiling of Single Formalin Fixed Tumour Sections: proteomics for translational research. Sci Rep 2016; 6: 34949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suomi T, Corthals GL, Nevalainen OS, et al. Using Peptide-Level Proteomics Data for Detecting Differentially Expressed Proteins. J Proteome Res 2015; 14: 4564–4570. [DOI] [PubMed] [Google Scholar]
- 26.Chen YH, Hung MC, Li LY. EZH2: a pivotal regulator in controlling cell differentiation. American journal of translational research 2012; 4: 364–375. [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang H, Vollmer M, De Geyter M, et al. Characterization of an immortalized human granulosa cell line (COV434). Mol Hum Reprod 2000; 6: 146–153. [DOI] [PubMed] [Google Scholar]
- 28.Chan-Penebre E, Armstrong K, Drew A, et al. Selective killing of SMARCA2- and SMARCA4-deficient small cell carcinoma of the ovary, hypercalcemic type cells by inhibition of EZH2: In vitro and in vivo preclinical models. Mol Cancer Ther 2017. [DOI] [PubMed] [Google Scholar]
- 29.Knutson SK, Warholic NM, Wigle TJ, et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc Natl Acad Sci U S A 2013; 110: 7922–7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knutson SK, Kawano S, Minoshima Y, et al. Selective inhibition of EZH2 by EPZ-6438 leads to potent antitumor activity in EZH2-mutant non-Hodgkin lymphoma. Mol Cancer Ther 2014; 13: 842–854. [DOI] [PubMed] [Google Scholar]
- 31.Poirier JT, Gardner EE, Connis N, et al. DNA methylation in small cell lung cancer defines distinct disease subtypes and correlates with high expression of EZH2. Oncogene 2015; 34: 5869–5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kotake Y, Cao R, Viatour P, et al. pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4alpha tumor suppressor gene. Genes Dev 2007; 21: 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Black MM, Kurdyla JT. Microtubule-associated proteins of neurons. J Cell Biol 1983; 97: 1020–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vidal M, Cusick ME, Barabasi AL. Interactome networks and human disease. Cell 2011; 144: 986–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mair B, Kubicek S, Nijman SM. Exploiting epigenetic vulnerabilities for cancer therapeutics. Trends Pharmacol Sci 2014; 35: 136–145. [DOI] [PubMed] [Google Scholar]
- 36.Bracken AP, Pasini D, Capra M, et al. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. EMBO J 2003; 22: 5323–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kadoch C, Williams RT, Calarco JP, et al. Dynamics of BAF-Polycomb complex opposition on heterochromatin in normal and oncogenic states. Nat Genet 2017; 49: 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stanton BZ, Hodges C, Calarco JP, et al. Smarca4 ATPase mutations disrupt direct eviction of PRC1 from chromatin. Nat Genet 2017; 49: 282–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sher F, Rossler R, Brouwer N, et al. Differentiation of neural stem cells into oligodendrocytes: involvement of the polycomb group protein Ezh2. Stem Cells 2008; 26: 2875–2883. [DOI] [PubMed] [Google Scholar]
- 40.Adhikary G, Grun D, Balasubramanian S, et al. Survival of skin cancer stem cells requires the Ezh2 polycomb group protein. Carcinogenesis 2015; 36: 800–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan J, Li B, Lin B, et al. EZH2 phosphorylation by JAK3 mediates a switch to noncanonical function in natural killer/T-cell lymphoma. Blood 2016; 128: 948–958. [DOI] [PubMed] [Google Scholar]
- 42.Chun HJ, Lim EL, Heravi-Moussavi A, et al. Genome-Wide Profiles of Extra-cranial Malignant Rhabdoid Tumors Reveal Heterogeneity and Dysregulated Developmental Pathways. Cancer Cell 2016; 29: 394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ueda K, Yoshimi A, Kagoya Y, et al. Inhibition of histone methyltransferase EZH2 depletes leukemia stem cell of mixed lineage leukemia fusion leukemia through upregulation of p16. Cancer Sci 2014; 105: 512–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suva ML, Riggi N, Janiszewska M, et al. EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Res 2009; 69: 9211–9218. [DOI] [PubMed] [Google Scholar]
- 45.Delbridge AR, Grabow S, Strasser A, et al. Thirty years of BCL-2: translating cell death discoveries into novel cancer therapies. Nat Rev Cancer 2016; 16: 99–109. [DOI] [PubMed] [Google Scholar]
- 46.Hubaux R, Thu KL, Coe BP, et al. EZH2 promotes E2F-driven SCLC tumorigenesis through modulation of apoptosis and cell-cycle regulation. J Thorac Oncol 2013; 8: 1102–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Galderisi U, Jori FP, Giordano A. Cell cycle regulation and neural differentiation. Oncogene 2003; 22: 5208–5219. [DOI] [PubMed] [Google Scholar]
- 48.Ng JM, Martinez D, Marsh ED, et al. Generation of a mouse model of atypical teratoid/rhabdoid tumor of the central nervous system through combined deletion of Snf5 and p53. Cancer Res 2015; 75: 4629–4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deodhar KK, Suryawanshi P, Shah M, et al. Immature teratoma of the ovary: a clinicopathological study of 28 cases. Indian J Pathol Microbiol 2011; 54: 730–735. [DOI] [PubMed] [Google Scholar]
- 50.Epizyme I. Epizyme Announces Updated Tazemetostat Phase 1 Data Showing Clinical Activity in a Broader Range of Adults With INI1-Negative and SMARCA4-Negative Solid Tumors. cited Apr 11, 2017 http://wwwepizymecom/wp-content/uploads/2015/09/EPZM_News_2015_9_26_General_Releasespdf.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Proteins identified by mass spectrometry in BIN67 cells treated with either DMSO or 1 uM EPZ-6438 for 7 d
Table S2. Differentially expressed proteins in BIN67 cells treated with EPZ-6438 versus DMSO
Table S3. Biological functions affected by EPZ-6438 treatment
Table S4. Proteins that contributed to the significantly altered biological functions by EPZ-6438 treatment
Figure S1. Effect of EZH2 depletion in ovarian cancer cell lines
Figure S2. Effect of EPZ-6438 on histone H3K27me3
Figure S3. Responses of ovarian cancer cell lines to EZH2 inhibitors in 9-day survival assays
Figure S4. EPZ-6438 suppressed clonogenic activity of COV434 cells
Figure S5. SMARCA4-deficient lung cancer cells and ovarian clear cell carcinoma cells are not sensitive to EPZ6438
Figure S6. Effect of EPZ-6438 in Bin67 xenograft models
Figure S7. Mouse body weight changes in BIN67 and SCCOHT-1 xenograft models for the EZH2 inhibitor efficacy studies
Figure S8. Unsupervised clustering analysis of proteins identified by mass spectrometry in BIN67 cells treated with DMSO or EPZ-6438 for 7 d (n=3)
Figure S9. Clustering analysis of proteins involved in each significantly altered biological function predicted by IPA analysis
Figure S10. Cytotoxic agents do not induce neuron-like morphologies in SCCOHT cells