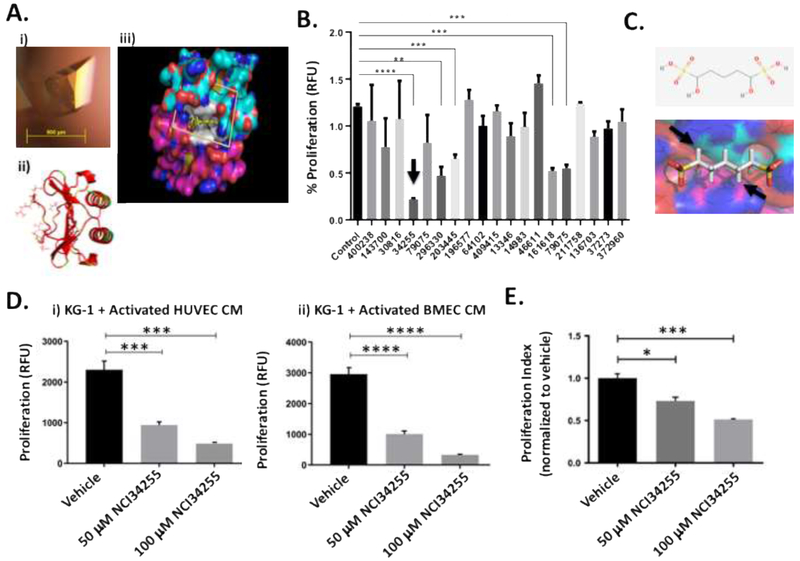
Figure 3. Identification of small molecule inhibitor of IL-8 that can significantly reduce leukemia cell expansion.
(A) Representative IL-8 crystal grown using the vapor diffusion hanging drop method. IL8 crystal yielding complete X-ray diffraction data set to 0.95 Å grown in 0.17 M Ammonium acetate, 0.085 M Sodium citrate tribasic dihydrate pH 5.6, 20% w/v Polyethylene glycol 4,000 and 15% v/v Glycerol (i). The IL-8 dimer is shown as a ribbon diagram in red. Side chains are shown for residues implicated in receptor binding based on mutagenesis studies (ii). The molecular surface of the IL-8 dimer is shown with blue for nitrogen, red for oxygen, cyan for carbon in the (top IL-8 subunit) and magenta for carbon (bottom IL-8 subunit). Residues implicated in receptor binding are shown in white. The scoring grid for molecular docking is shown as a box. 139,735 NCI/DTP compounds were docked into the binding site in silico to predict the binding affinity for IL-8 (iii). (B) Docking studies identified 19 small molecules capable of binding IL-8’s receptor binding site. Treatment of KG-1 cells with these molecules identified 5 that were able to significantly decrease KG-1 proliferation. The best hit molecule is identified with an arrow. ** p < 0.01; *** p < 0.001; **** p < 0.0001 (C) The best hit molecule was NCI34255 which interferes with binding at the homodimeric interface of two IL-8 sub-units (see arrows). (D) KG-1 cells were grown in CM from KG-1 activated ECs (HUVECs and BMECs) supplemented with NCI34255. The presence of NCI34255 was able to significantly decrease IL-8 induced cell proliferation contrary to the enhanced proliferation observed in previous CM studies. *** p < 0.001; **** p < 0.0001 (E) Supplementation of co-cultures comprising KG-1/BMECs with NCI34255 showed significant decreases in non-adherent KG-1 cell proliferation indicating the ability of NCI34255 to overcome activated EC-generated IL-8 signaling. * p < 0.05; *** p < 0.001