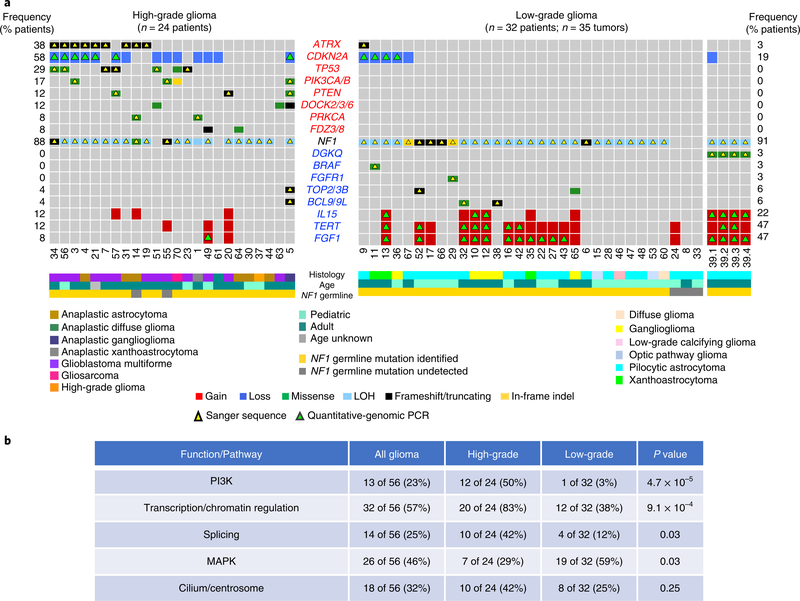
Fig. 2 |. Landscape of somatic genomic alterations in NF1-glioma.
a, Integrated matrix of 59 glioma samples from 56 patients and gene variants (SNVs, indels, and significant CNVs) observed in NF1-glioma (left panel, high-grade glioma; right panels, low-grade glioma). Rows and columns represent genes and tumor samples, respectively. Genomic alterations, age, the histology of glioma, and NF1 germline mutations are indicated. NFI-glioma samples are sorted by their mutation profiles, except for patient no. 5, hypermutated high-grade glioma, and patient no. 39, including four spatially distinct glioma samples, which are shown at the last columns of left and right panel, respectively. Recurrently mutated genes are selected for their previously established association with glioma (ATRX, CDKN2A, TP53, PIK3CA/B, PTEN, BRAF, FGFR1 and FGF1, PRKCA, TERT), cancer biology (DOCK2/3/6, FDZ3/8, BCL9/9L, TOP2/3B), and immune functions (IL15, DGKQ). Genes are sorted according to higher frequency (percentage of patients) in high-grade (top, red) or low-grade gliomas (bottom, blue), respectively. Validations by Sanger sequencing (SNVs) and quantitative-genomic PCR (gains and losses) are indicated by yellow and green triangles, respectively. LOH, loss of heterozygosity. b, Function/pathway analysis of damaging somatic mutations and CNVs. Genetic alterations in NF1-gliomas grouped into PI3K, transcription/chromatin regulation, splicing, MAPK, and cilium/centrosome functions. A significantly higher frequency of genetic alterations in PI3K, transcription/chromatin regulation, and splicing pathway were observed in high-grade glioma (n = 24; P = 4.7×10−5, P = 9.1×10−4, and P = 0.03, respectively; two-sided Fisher’s exact test), while mutations in the MAPK pathway were more frequent in low-grade glioma (n = 32; P = 0.03, two-sided Fisher’s exact test). The integrated matrices of NF1-glioma and gene pathway alterations are reported in Extended Data Figs. 9 and 10.