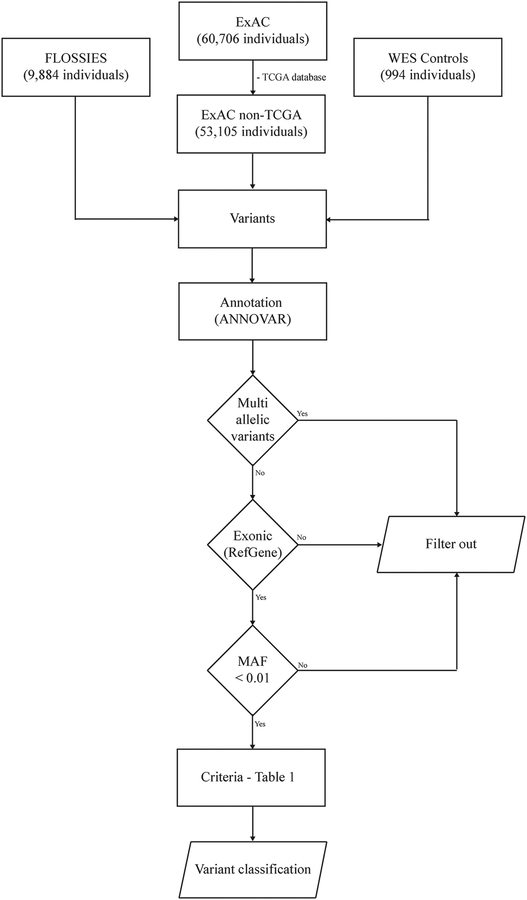
Figure 1 -. Flowchart with a summary of parameters used for variant selection and filtering.
Our analysis is based on a pooled dataset composed by 63,983 individuals, after excluding the TCGA dataset from the ExAC consortium. Variants were annotated using ANNOVAR and selected based on gene region equivalent to exonic and with MAF < 0.01. The canonical transcript NM_00546.5 was used as sequence reference for gene region location based on the RefGene database. Variant classification was established by an original algorithm based on the REVEL score, clinical significance evidences, and the impact on transcriptional activity. Abbreviations: TCGA, The Cancer Genome Atlas; ExAC, Exome Aggregation Consortium; FLOSSIES, Fabulous Ladies Over Seventy; WES, Whole-Exome Sequencing; MAF, Minor Allele Frequency.