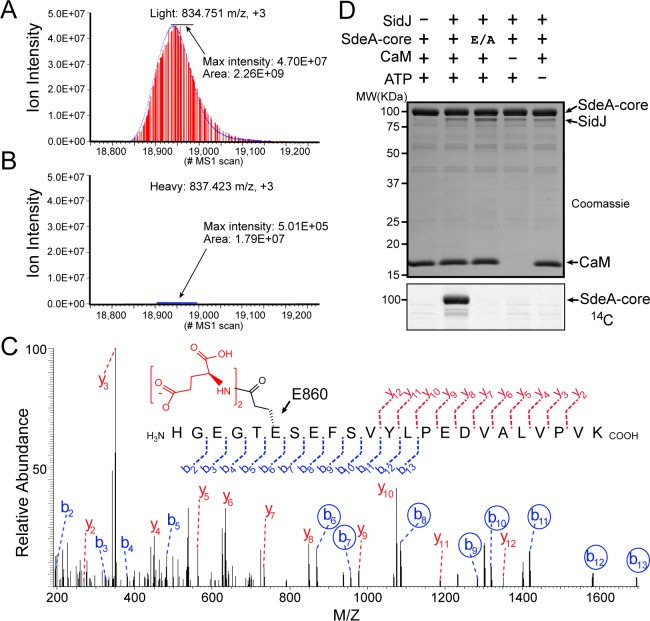
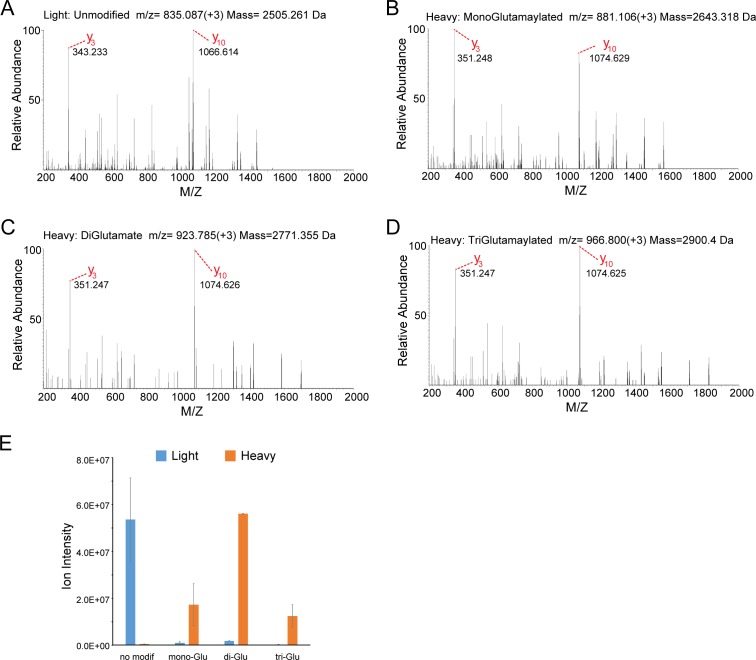
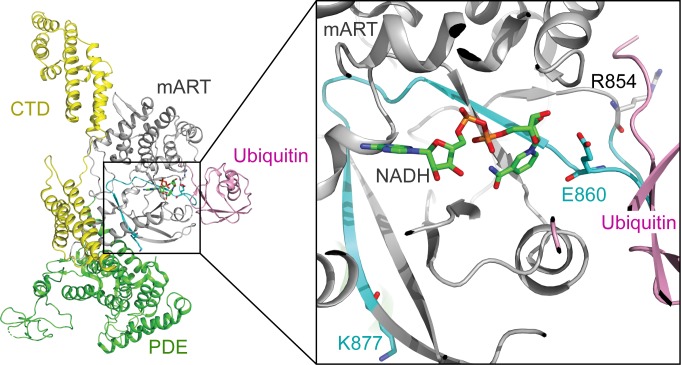
Figure 4. SidJ catalyzes the polyglutamylation of SdeA.
Reconstructed ion chromatograms for the SdeA peptide (residues 855–877) from (A) cells grown in light medium and co-transfected with GFP-SdeA and mCherry vector control or (B) cells grown in heavy medium and co-transfected with GFP-SdeA and mCherry-SidJ. (C) MS2 spectrum of a di-glutamylated SdeA mART peptide with b ions labeled in blue and y ions labeled in red. The peptide sequence corresponding to fragmentation is depicted above the spectrum. Circled b ions represent a mass increase corresponding to diglutamylation (258.085 Da). (D) In vitro glutamylation of SdeA with [U-14C]glutamate. E/A corresponds to the E860A point mutant of SdeA. Proteins were separated by SDS-PAGE and visualized with Coomassie stain (top panel). An autoradiogram of the gel is shown in the bottom panel.