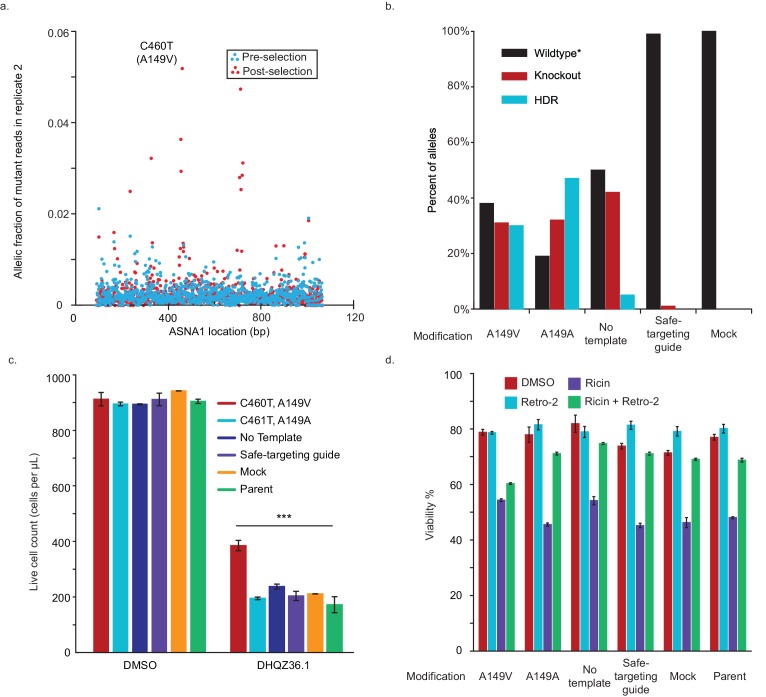
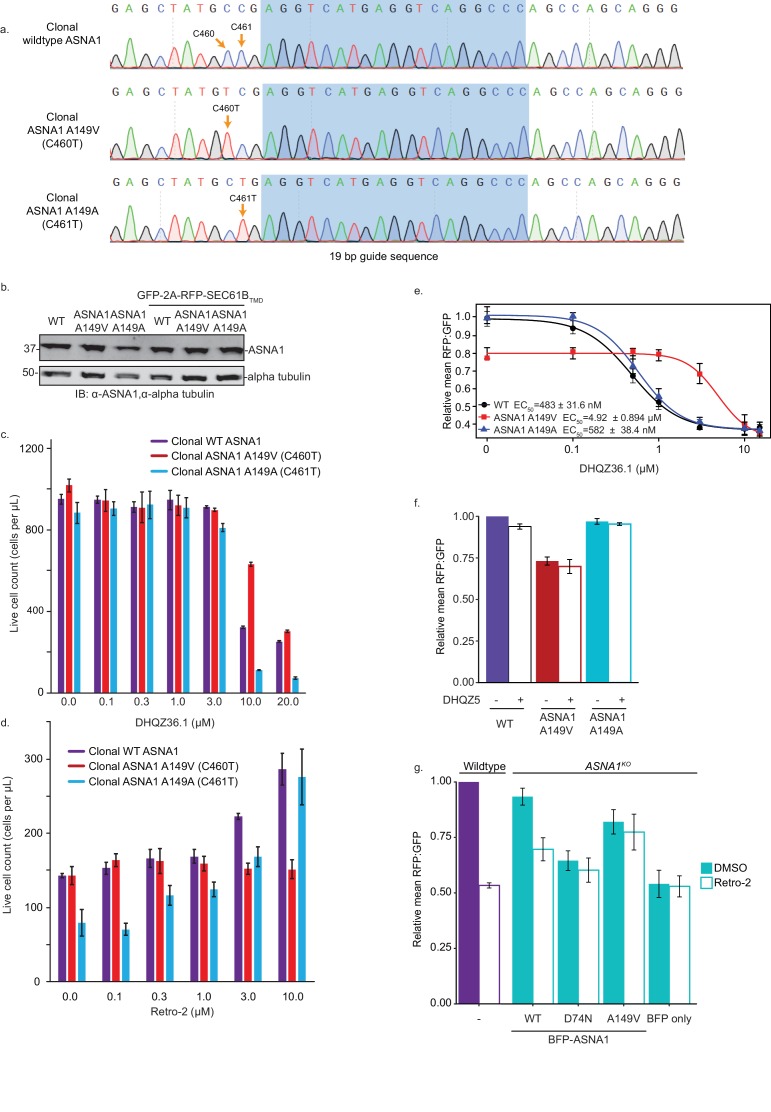
Figure 4. Isolation and characterization of A149V ASNA1, a Retro-2-resistance allele.
(A) Schematic of ASNA1 mutagenesis by CRISPR-X. A 172 sgRNA library tiling the ASNA1 coding region was lentivirally infected in a K562 cell line expressing a dCas9-AID*Δ N-terminal fusion. The pool was then grown in duplicate in the presence of 20 μM DHQZ36.1 for 4 weeks. ASNA1 was amplified separately from cDNA of naive and treated cell populations before being sequenced by Nextera XT. (B) Plot of the frequency of ASNA1 alleles across the gene-body (x-axis) in the initial CRISPR-X population (pre-selection) and in one replicate of the selected population. Allelic fraction was calculated by determining the per-base variant frequency that is the number of reads which contain a mutated base at a given position vs the number of reads which contain the wildtype base at that position. Bases which had less than 500× read coverage were excluded, resulting in no resolution of the 5’/3’ ends of ASNA1. The top selected mutation in both replicates is highlighted. (C) A homology model of human ASNA1 (SWISS-MODEL: O43681) with the location of the top mutated residue, A149, enriched after selection by DHQZ36.1 toxicity highlighted in red on one ASNA1 (colored in green) of the homodimer. (D) A149V was installed by homology directed repair in a K562 cell line expressing Cas9. The resulting mutant line was treated with ricin toxin in the presence of 10 μM Retro-2 or DMSO. Live cells were counted using forward/side scatter by cytometry. Shown are bar graphs of the mean with standard error from three technical replicates. Also shown are data for five control lines treated in the same way. See Materials and methods for further details. (E) Clonal wildtype (black circles), ASNA1 A149V (C460T) (red squares), and ASNA1 A149A (C461T) (blue triangles) K562 cell lines with the GFP-2A-RFP-SEC61BTMD reporter were pre-treated with Retro-2 with the indicated concentrations for 1 hr prior to induction with dox for approximately 18 hr and FACS analysis. Shown are the dose-response curves for the reporter RFP to GFP ratios as relative means (three experiments) to mock-treated wildtype cells. The dose response was modeled using the four-parameter logistic regression to determine the half maximal effective concentration (EC50 ± standard error). Error bars for the means represent the standard error calculated from the four-parameter logistic regression.