Figure 1.
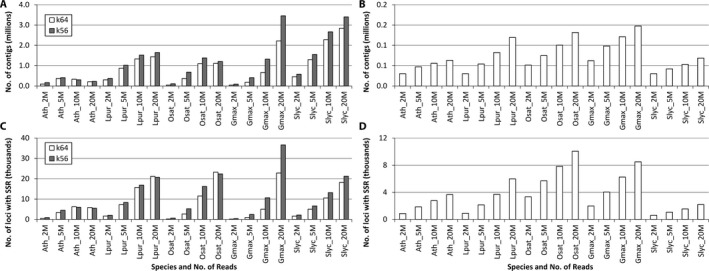
Number of contigs (A, B) and number of loci with simple sequence repeats (C, D) derived from the genome (A, C) and transcriptome (B, D) assemblies. Two k‐mer sizes (k56 and k64) were used in the genome assemblies. Species analyzed are abbreviated as follows: Ath = Arabidopsis thaliana (Arabidopsis); Lpur = Lablab purpureus (lablab); Osat = Oryza sativa (rice); Gmax = Glycine max (soybean); Slyc = Solanum lycopersicum (tomato). Four different depths of sequencing were used (2M, 5M, 10M, and 20M reads). Further metrics are given in Appendices S1 and S3.
