Abstract
With recent advances in DNA sequencing technologies, fast acquisition of large-scale genomic data has become commonplace. For cancer studies, in particular, there is an increasing need for the classification of cancer type based on somatic alterations detected from sequencing analyses. However, the ever-increasing size and complexity of the data make the classification task extremely challenging. In this study, we evaluate the contributions of various input features, such as mutation profiles, mutation rates, mutation spectra and signatures, and somatic copy number alterations that can be derived from genomic data, and further utilize them for accurate cancer type classification. We introduce a novel ensemble of machine learning classifiers, called CPEM (Cancer Predictor using an Ensemble Model), which is tested on 7,002 samples representing over 31 different cancer types collected from The Cancer Genome Atlas (TCGA) database. We first systematically examined the impact of the input features. Features known to be associated with specific cancers had relatively high importance in our initial prediction model. We further investigated various machine learning classifiers and feature selection methods to derive the ensemble-based cancer type prediction model achieving up to 84% classification accuracy in the nested 10-fold cross-validation. Finally, we narrowed down the target cancers to the six most common types and achieved up to 94% accuracy.
Subject terms: Cancer genomics, Machine learning
Introduction
Cancer is a complex disease that refers to the phenomenon of abnormal cellular proliferation, invasion, or metastasis in human tissues and blood. It is caused by the acquisition of a series of genomic alterations. Hence, an understanding of the genetic characteristics of cancer is crucial for accurate diagnosis and treatment. Next-generation sequencing techniques have recently been widely applied to cancer research to characterize various different types of genomic alterations in cancer genomes1–4. Large-scale cancer genome studies have revealed that the patterns of genomic alterations are often cancer type-specific. Lawrence et al. found that cancer mutations vary across cancer types, with genes mutated in a cancer-specific manner5. Hoadley et al. also confirmed that tissue-specific genomic features in cancers are the dominant signals for classifying cancer subtypes6. In addition, somatic copy number alterations and mutation spectra also display tissue-specific patterns7–9.
Using the tissue-specific nature of somatic alterations in cancer, a number of prediction methods for cancer type were recently developed by employing machine learning classifiers and various mutation features to improve classification accuracy. Marquard et al. used a random forest classifier (i.e., one-vs-rest (OvR) binary classifiers for multiclass classification) with a feature set consisting of somatic point mutations of known cancer-associated genes, mutation frequencies, and copy number profiles from the Catalogue of Somatic Mutations in Cancer (COSMIC) database10 to identify tissues of origin11. The authors reported up to 85% maximum accuracy across six cancer types and 69% across 10 cancer types. Chen et al. used a support vector machine (SVM) classifier with official gene symbols, mutations, chromosome, and pathways as feature sets, which led to the maximum average accuracy of 62% across 17 cancer types12. Yuan et al. introduced a novel clustering-based feature selection scheme with a deep neural network classifier (i.e., multilayer perceptron) for cancer classification13. In their work, only somatic point mutations were used for input feature data, which led to the maximum accuracy of 64% across 12 cancer types. However, these previous studies have not fully assessed and exploited biologically meaningful input features and advanced machine learning techniques. Furthermore, most of them were able to handle only a small subset of the cancer types from the database for classification.
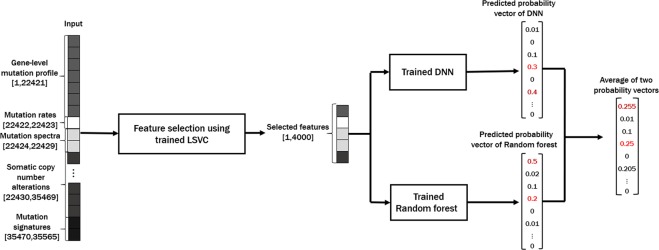
To address these problems, we propose a novel cancer type classification method, the Cancer Predictor using an Ensemble Model (CPEM) (see Fig. 1), which is based on the combination of advanced machine learning algorithms with various types of cancer somatic alterations and their derived features as input. To identify which input features are useful in classification, we conducted a comprehensive study on how various genetic alterations, such as gene-level mutation profiles, mutation rates, mutation spectra and signatures, and gene-level copy number alterations affect the accuracy of the classifier and we discuss the biological reasoning behind this result. We further investigated the performance of various state-of-the-art machine learning classifiers, both for feature selection and for cancer classification, and developed a novel cancer type classifier based on an ensemble of deep neural network (DNN) and random forest classifiers. CPEM can classify a large number of cancer types with high accuracy (i.e., the average accuracy of 84% across 31 cancer types in the nested 10-fold cross-validation). When we focused on the six most common cancer types, we were able to achieve up to 94% average accuracy. To the best of our knowledge, the proposed method is the most accurate multiclass cancer type classification method yet-devised that is able to predict the largest class of cancer types, covering the entire cancer list in the TCGA database3.
Figure 1.
Overview of the CPEM cancer type prediction workflow. The first step in our workflow is building a set of feature vectors from the TCGA database. The feature vector consists of 22,421 gene-level mutation profiles, two mutation rates, six mutation spectra, 13,040 gene-level copy number alterations, and 96 mutation signatures. The next step is the feature vector dimension reduction by linear support vector classifier (LSVC)-based feature selection. In the final step, the two machine learning classifiers which were trained by the same selected feature set (a deep neural network and a random forest) are combined to build an ensemble model for the final cancer type prediction.
Results
Experimental setup
All experiments were conducted using a workstation equipped with an Intel Xeon CPU E5-2640 CPU with four NVIDIA GTX 1080Ti GPUs. We used TensorFlow (version 1.9.0) to implement the proposed deep neural network. Our DNN architecture is a multi-layer perceptron with a multinomial class cross-entropy with a softmax loss function. We used the Rectified Linear Unit as the activation function.
Selecting the proper number of parameters and hidden layers (i.e., tuning hyperparameters) of the DNN is important for better prediction accuracy and faster training time. We empirically chose our DNN with three hidden layers where each layer has 2048 parameters. To train the neural network, we used the Adam optimizer14, a commonly used stochastic optimization algorithm for machine learning, with a learning rate of 10−5 and 40% dropout per iteration to prevent overfitting. Other machine learning classifiers were implemented using the scikit-learn library15 in Python 3.6.1.
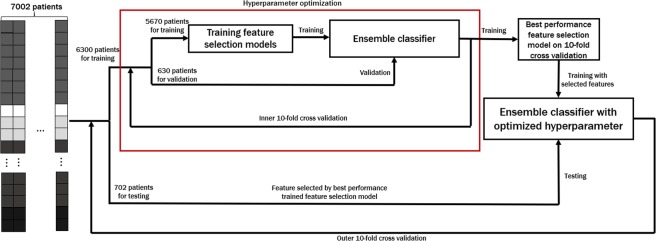
As shown in Fig. 2, the optimal number of input features and the feature selection method are chosen to maximize the accuracy over inner 10-fold cross-validation. Once feature selection is finalized, we tested the performance of CPEM using the independent testing data that was not a part of the training data.
Figure 2.
Overview of the training and testing process of CPEM. We employ nested 10-fold cross-validation where the optimal feature selection is performed using the inner 10-fold cross-validation while the accuracy of the proposed method is evaluated using the outer 10-fold cross-validation. In the training step, two machine learning classifiers are trained with the same selected feature set by optimal feature selection. Note that we use the testing data that is not used in the feature selection optimization step in every iteration of the outer cross-validation.
Efficacy of Various Mutation Features
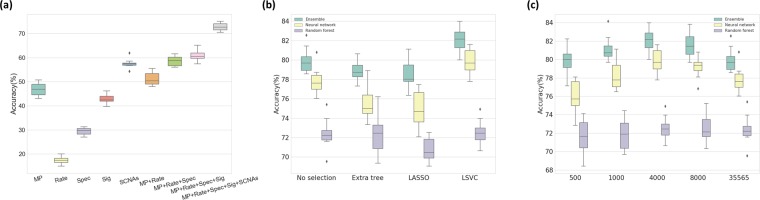
To assess how each class of input feature affected the classification accuracy, we used a random forest classifier as an initial prediction model and measured changes in classification performance as each feature was added using 10-fold cross-validation. Starting from the gene-level mutation profiles only, we consecutively added other feature groups, including mutation rates, mutation spectra, gene-level SCNAs, and mutation signatures, and measured the classification accuracy for each time. The accuracy for each feature group was 46.9%, 51.2%, 58.5%, 61.0%, and 72.7% respectively (Fig. 3a).
Figure 3.
(a) Classification accuracy of a random forest classifier on each combination of feature groups. MP: Gene-level Mutation Profiles, Rate: Mutation Rates, Spec.: Mutation Spectra, SCNAs: Somatic Copy Number Alterations, Sig.: Mutation Signatures. (b) Comparison of feature selection methods. The boxplot shows the classifier accuracy measured by the inner 10-fold cross-validation for each feature selection method. It is shown that LSVC feature selection performs best on all three classifiers. (c) Classification accuracy for various feature sizes generated by the LSVC method. It is shown that the classification accuracy reaches its peak performance at around 4,000 to 8,000 features measured to optimize the DNN and random forest by the inner 10-fold cross-validation.
To further deduce the features that most contribute to the accuracy of the classification, we selected the top ten features based on the importance score from the classifier. These features included two mutation rates, one mutation signature (CCT.C > T), two mutation spectra (C > T, C > A) and five mutated genes [von Hippel-Lindau tumor suppressor (VHL); isocitrate dehydrogenase (NADP(+)) 1, cytosolic (IDH1); B-Raf proto-oncogene, serine/threonine kinase (BRAF); APC, WNT signaling pathway regulator (APC); KRAS proto-oncogene, GTPase (KRAS)] (Supplementary Fig. S1). This result is consistent with previous studies that identified distinct mutational landscapes from many different types of cancer genomes8,9,16. For example, BRAF is involved in intracellular signaling associated with cell growth induction and is frequently mutated in some human cancers17. Acquiring mutations in KRAS is an essential step in the pathogenesis of many cancers18. This result confirms that genetic features play an important role in cancer initiation and progression, and also contribute to improving the accuracy of cancer classification. The biological meaning of this result will be discussed further in the ‘Discussion’.
Optimal Feature Selection
To choose the best feature selection method for our data, we tested the three most commonly used supervised feature selection algorithms—the extra tree-based, LASSO, and LSVM approaches. In this experiment, each feature selection method was applied to the same input feature set to reduce its size by 90% (i.e., only 10% of the input feature will be retained after selection), and the prediction accuracy of different classifiers using selected features was measured.
The accuracy of the three classifiers (i.e., ensemble, neural network, random forest) on the data processed by the three different supervised feature selection methods was compared (Fig. 3b; the inner 10-fold cross-validation results are shown using a boxplot). In general, the LASSO and LSVC selection methods performed well for the neural network classifier, and the extra tree-based selection method worked well for the random forest classifier. This is because LASSO and LSVC use the linear classifier similar to the perceptron model in the neural network, and extra tree-based selection is similar to the decision trees in a random forest. Of the LASSO and LSVC methods, LSVC displayed higher classification accuracy.
The performance of the classifier is affected by the selection method and by the number of selected features (i.e., the amount of feature dimension reduction). Figure 3c summarizes the classification accuracy for the different number of selected features, measured using the inner 10-fold cross-validation. For 35,565 features, approximately 4,000 to 8,000 selected features showed the best result, which is about 10% to 20% of the original feature set. By doing so, a higher accuracy could be achieved, up to a 2.34% increase in the outer 10-fold cross-validation, with less training time due to the small training set size.
Putting it all Together: CPEM Results
Based on the feature selection scheme and the optimal number of features found in the previous steps, we construct the proposed ensemble-based cancer prediction model, CPEM. We have tested various combinations of classifiers and finally chosen a deep neural network and a random forest for our ensemble model. By combining two classifiers, we have reached up to 84% average accuracy on the testing data for 31 cancer types. This result is about 6 and 11 percentage points higher than the accuracy of conventional machine learning classifiers, such as fully-connected deep neural networks and random forests, respectively. Figure 4 shows the result of the outer 10-fold cross-validation for CPEM and the other classifiers, and Table 1 shows the detailed description of per-cancer type experimental result.
Figure 4.
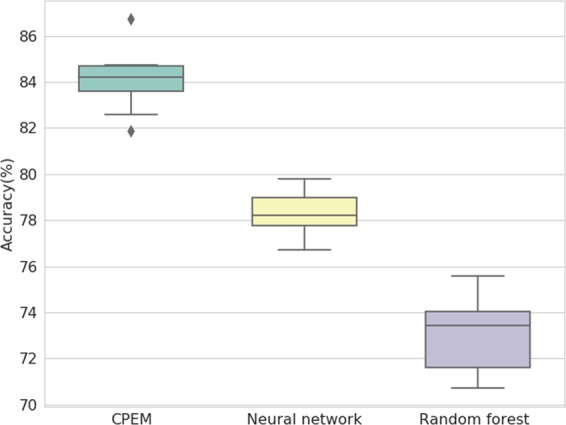
Comparison of CPEM and other conventional machine learning classifiers. The boxplot shows the accuracy measured by the outer 10-fold cross-validation for each machine learning model after optimization through inner 10-fold cross-validation. The average classification accuracy of CPEM is about 6 and 11 percentage points higher than those of a fully-connected deep neural network and a random forest, respectively.
Table 1.
Cancer type, sample size, number of mutated genes, number of copy number-altered genes, average precision, average recall, average F1 score and average classification accuracy of 31 cancer types used in our experiment.
| Cancer type | Sample size | # of mutated genes | # of copy number altered genes | Precision (%) | Recall (%) | F1 score (%) | Accuracy (%) |
|---|---|---|---|---|---|---|---|
| Adrenocortical carcinoma (ACC) | 88 | 6,904 | 12,725 | 96.10 | 84.09 | 89.70 | 84.09 |
| Bladder urothelial carcinoma (BLCA) | 127 | 11,674 | 11,223 | 92.04 | 63.78 | 75.35 | 62.20 |
| Breast invasive carcinoma (BRCA) | 967 | 15,716 | 12,178 | 83.19 | 92.14 | 87.44 | 92.86 |
| Cervical and endocervical cancers (CESC) | 191 | 12,165 | 11,505 | 69.63 | 68.06 | 68.84 | 71.89 |
| Cholangiocarcinoma (CHOL) | 35 | 3,511 | 9,707 | 73.68 | 40.00 | 51.85 | 31.43 |
| Colorectal adenocarcinoma (COADREAD) | 220 | 14,941 | 11,509 | 87.73 | 87.73 | 87.73 | 88.18 |
| Lymphoid Neoplasm Diffuse Large B-cell Lymphoma (DLBC) | 48 | 6,457 | 9,475 | 90.91 | 62.50 | 74.07 | 58.33 |
| Esophageal carcinoma (ESCA) | 184 | 13,557 | 1,1534 | 81.18 | 58.70 | 68.35 | 63.04 |
| Glioblastoma multiforme (GBM) | 280 | 8,069 | 11,410 | 81.85 | 88.57 | 85.08 | 89.64 |
| Head and Neck squamous cell carcinoma (HNSC) | 279 | 12,783 | 11,270 | 70.65 | 74.19 | 72.38 | 77.42 |
| Kidney Chromophobe (KICH) | 66 | 3,698 | 10,431 | 96.49 | 83.33 | 89.43 | 83.33 |
| Kidney renal clear cell carcinoma (KIRC) | 410 | 9,900 | 11,562 | 88.36 | 94.39 | 91.27 | 94.39 |
| Kidney renal papillary cell carcinoma (KIRP) | 161 | 7,332 | 10,568 | 83.23 | 80.12 | 81.65 | 78.88 |
| Acute Myeloid Leukemia (LAML) | 182 | 1,369 | 7,719 | 84.50 | 92.86 | 88.48 | 93.41 |
| Brain Lower Grade Glioma (LGG) | 280 | 4,732 | 10,190 | 89.88 | 82.50 | 86.03 | 82.86 |
| Liver hepatocellular carcinoma (LIHC) | 193 | 9,985 | 11,980 | 81.59 | 68.91 | 74.72 | 66.84 |
| Lung adenocarcinoma (LUAD) | 230 | 13,931 | 11,071 | 86.51 | 80.87 | 83.60 | 79.57 |
| Lung squamous cell carcinoma (LUSC) | 177 | 13,487 | 11,212 | 73.33 | 74.58 | 73.95 | 78.53 |
| Ovarian serous cystadenocarcinoma (OV) | 311 | 8,435 | 11,684 | 81.69 | 90.35 | 85.80 | 91.96 |
| Pancreatic adenocarcinoma (PAAD) | 149 | 10,144 | 8,744 | 83.23 | 89.93 | 86.45 | 89.93 |
| Pheochromocytoma and Paraganglioma (PCPG) | 166 | 2,125 | 10,103 | 86.78 | 90.96 | 88.82 | 89.16 |
| Prostate adenocarcinoma (PRAD) | 331 | 6,116 | 9,996 | 80.16 | 90.33 | 84.94 | 87.61 |
| Sarcoma (SARC) | 243 | 7,416 | 12,099 | 86.00 | 70.78 | 77.65 | 69.14 |
| Skin Cutaneous Melanoma (SKCM) | 341 | 17,085 | 11,883 | 90.05 | 92.67 | 91.54 | 92.67 |
| Stomach adenocarcinoma (STAD) | 286 | 16,607 | 11,745 | 76.09 | 79.02 | 77.53 | 80.77 |
| Testicular Germ Cell Tumors (TGCT) | 155 | 5,904 | 9,133 | 91.02 | 98.07 | 94.41 | 98.71 |
| Thyroid carcinoma (THCA) | 403 | 3,910 | 8,721 | 85.14 | 93.80 | 89.26 | 93.80 |
| Thymoma (THYM) | 121 | 1,805 | 9,305 | 86.14 | 71.90 | 78.38 | 76.03 |
| Uterine Corpus Endometrial Carcinoma (UCEC) | 242 | 18,412 | 12,133 | 81.39 | 77.69 | 79.49 | 76.86 |
| Uterine Carcinosarcoma (UCS) | 56 | 5,508 | 11,611 | 67.74 | 37.50 | 48.28 | 35.71 |
| Uveal Melanoma (UVM) | 80 | 1,340 | 9,559 | 80.00 | 85.33 | 82.58 | 83.75 |
| Total | 7,002 | 22,421 | 13,040 | 83.43 | 78.89 | 80.49 | 84.09 |
For better understandings of CPEM, we tested four well-known machine learning classifiers and further analyzed the correlation of the methods with each other. Table 2 shows the classification results of widely used classifiers, including DNN, OvR SVM, random forest, and k-nearest neighbors (KNN) clustering classifiers, in a pairwise fashion. In this table, A ∪ B is the probability that either A or B classifiers correctly predict the cancer type (total correct prediction), A ∩ B is the probability that both A and B classifiers correctly predict the cancer type (common correct prediction), and A−B and B−A are the probability that only either A or B predicts the result correctly. The classifiers were less correlated when A ∩ B was low and A−B and B−A were high. In addition, since we listed A and B in the table in descending order, it indicates that A is the better predictor (i.e., prediction accuracy is higher), so that B−A is the upper bound of the accuracy gain when the ensemble method is used. It is also important that the total correct prediction rate should be high (A ∪ B), and each classifier should predict higher than 50% accuracy (otherwise, it will impair the performance of the ensemble). In our experiment, we observed that the KNN clustering classifier performed worst, with only 48.59% prediction accuracy. It does not improve the accuracy when combined with other methods via ensemble. The random forest and OvR SVM performed similarly, with a respective prediction accuracy of 73.79% and 72.85%. Our DNN classifier outperformed all other classifiers, with a prediction accuracy of 82.25% which was approximately 10% higher than that of a random forest. The random forest classifier was slightly more accurate than the OvR SVM and was less correlated with DNN (i.e., lower A ∩ B). Therefore, the ensemble model combining the DNN and the random forest were the best choice. CPEM increased the accuracy of the DNN by 2%, which led to an average prediction accuracy of 84.09% for the 31 cancer types.
Table 2.
Classification accuracy of various machine learning classifiers and their combinations with LSVC feature selection in the outer 10-fold cross validation.
| A | B | A ∩ B | A ∪ B | A−B | B−A | Ensemble (A, B) | |
|---|---|---|---|---|---|---|---|
| DNN (A), Random forest (B) | 82.25 | 73.79 | 67.81 | 88.23 | 14.44 | 5.98 | 84.09 |
| DNN (A), OvR SVM (B) | 82.25 | 72.85 | 70.21 | 84.89 | 12.04 | 2.64 | 80.95 |
| DNN (A), KNN (B) | 82.25 | 48.59 | 46.30 | 84.53 | 35.95 | 2.29 | 78.81 |
| Random forest (A), OvR SVM (B) | 73.79 | 72.85 | 62.58 | 84.06 | 11.21 | 10.27 | 78.79 |
| Random forest (A), KNN (B) | 73.79 | 48.59 | 43.87 | 78.51 | 29.92 | 4.71 | 67.30 |
| OvR SVM (A), KNN (B) | 72.85 | 48.59 | 44.50 | 76.94 | 28.35 | 4.08 | 68.41 |
This result confirms that the ensemble of a deep neural network and a random forest performs best.
We also compared our method to existing machine learning-based cancer type classification methods. Since it is not practically feasible to use exactly the same training data used in other studies, we collected the data of the same cancer types used in other studies from our database and compared the accuracy reported in the literature (Table 3). For the cancer types used in TumorTracer11, we achieved up to 91.42% accuracy for six types (85% in TumorTracer) and 90.28% accuracy for ten types (69% in TumorTracer) (Supplementary Figs S4 and S5). When using the same 12 cancer types used in DeepGene13, we were able to achieve up to 84.66% accuracy, which was 20 percentage points higher than that of DeepGene (64%) (Supplementary Fig. S6).
Table 3.
Comparison between CPEM and previously reported cancer type classification methods.
| # of samples | # of features | # of cancer types | Precision | Recall | F1 score | Accuracy | |
|---|---|---|---|---|---|---|---|
| DeepGene13 | 3122 | 1200 | 12 | N/A | N/A | N/A | 66.50 |
| *TumorTracer11 | 2820 | 232 | 6 | 85.83 | 84.95 | 85.39 | 85.00 |
| 4975 | 560 | 10 | 72.23 | 68.98 | 70.57 | 69.00 | |
| Chen et al.12 | 6751 | 101176 | 18 | 65.24 | 62.26 | 63.72 | 62.00 |
| *CPEM | 2763 | 4000 | 6 | 84.77 | 90.61 | 87.59 | 94.14 |
| 4823 | 4000 | 14 | 83.48 | 85.36 | 84.41 | 87.02 | |
| 7002 | 4000 | 31 | 83.43 | 78.89 | 80.49 | 84.06 |
Discussion
We constructed various feature groups from the collected data and confirmed a maximum of 72.2% accuracy through a combination of features using an initial prediction model based on a random forest. Feature importance was calculated to identify elements with a high impact in a combination of features with maximum accuracy. The top ten features with the highest importance were , CCT.C > T mutation signature, VHL mutation, IDH1 mutation, C > T mutation frequency, C > A mutation frequency, BRAF mutation, , APC mutation, and KRAS mutation (Supplementary Fig. S1). The frequency of mutation varies widely among samples according to the cancer type, ranging from 0.1 (pediatric cancer) to 100 (lung cancer) per Mb. High mutation frequencies are due to extensive exposure to well-known carcinogens, such as tobacco smoke (C > A mutation) and ultraviolet radiation (C > T mutation)9. We referred to the COSMIC10 data to identify signatures with a high frequency of CCT.C > T mutations, and confirmed that the frequency of CCT.C > T mutations was the highest in Signature 23 and 19. Signature 23 is present only in liver cancer samples, and Signature 19 is specific to pilocytic astrocytoma. According to the COSMIC data, VHL displayed mutation rates of 39% and 33% in liver cancer and paratesticular tissues, respectively. VHL is a transcription factor that plays a central role in the regulation of gene expression by oxygen and is involved in ubiquitination and degradation of hypoxia-inducible-factor19. IDH1 catalyzes the conversion of isocitrate to alpha-ketoglutarate (aKG) in normal conditions. The cancer-associated IDH1 mutation converts aKG to 2-hydroxyglutarate (2HG)20. In gliomas and malignancies, IDH1 mutations induce postmenopausal changes and promote tumorigenesis21. The frequency of IDH1 mutations in the COSMIC data is highest in the central nervous system (34%). BRAF is a crucial regulator of the extracellular signal-regulated kinase – mitogen activated protein kinase signaling pathway, leading to cell proliferation, differentiation, and survival. BRAF mutations are present at a high frequency (8%) in various cancers, particularly in melanomas (50%)22. APC has a wide range of functions from the regulation of the WNT signaling pathway to cell migration, apoptosis, and chromosome segregation23. APC mutations occur in 42% of colorectal cancer and 14% of small bowel cancer. KRAS is a well-known oncogene that is commonly found in pancreatic, colon, and lung cancers. KRAS induces tumorigenesis, regulates cell degeneration, and induces genomic instability.
Even though we already achieved KG high classification average accuracy of up to 84% for 31 cancer types from the TCGA database using our ensemble method, the classification accuracy could be increased further by focusing on cancer types with a sufficient number of samples. One observation we made is that the size of the training data is not evenly distributed across the cancer types in the database. Some cancer types have a very small number of training sets, which affects the performance of machine learning (Table 1). For example, we only have 35 samples for the CHOL cancer type, which resulted in low average accuracy of 31.43%. To circumvent this issue, we selected subsets of cancer types based on the number of samples. For cancer types with 200 or more samples, we were able to collect 14 cancer types from 4,823 samples and achieved an accuracy of up to 87%, which was about 4 percent points higher than using all 31 cancer types (Supplementary Fig. S2). If we increased the threshold to 300 or more samples, then six most frequent cancer types remained (BRCA, KIRC, OV, PRAD, SKCM, and THCA), and the classification accuracy further improved to 94%, which was about 10 percentage points higher than using all 31 cancer types (Supplementary Fig. S3).
To further demonstrate the wide applicability of the proposed method to other data (i.e., non-TCGA data), we trained and tested CPEM using the ICGC (International Cancer Genome Consortium) dataset, which provides sequencing data for 76 cancer projects. We collected somatic SNV and CNV data of 48 cancer projects performed on at least 10 samples. In this experiment, we used the same nested 10-fold cross-validation used for TCGA data to assess the performance. CPEM achieved up to 82.40% accuracy, while the accuracy of neural network and random forest are 77.93% and 74.83% in outer 10-fold cross-validation, respectively. This demonstrates that CPEM is effective on non-TCGA data as well.
We observed that, even though CPEM demonstrates superior performance on TCGA or ICGC only, its cross-platform application is not working well, which is somehow expected in a data-driven approach. For this experiment, we collected 124 BRCA, 265 ESCA, 428 PAAD and 451 BRCA samples from the non-TCGA ICGC data. Then we directly applied our CPEM trained using only TCGA data to these non-TCGA data, which resulted in the classification accuracy of 78.23%, 6.79%, 64.95% and 49.22% for PRAD, ESCA, PAAD and BRCA, respectively. We believe the reason for the poor performance on ESCA and BRCA is due to the platform inhomogeneity. According to the ICGC portal web page (https://icgc.org/node/70708), this non-TCGA ESCA data generated by Cancer Research UK seem to also include Barrett’s esophagus as well as esophageal cancer. We believe that the histological difference between Barrett’s esophagus and esophageal cancer is significant, and this is the reason why the TCGA-trained model showed poor performance in predicting non-TCGA ESCA. As for BRCA, we compared histopathological subtypes for the tumor samples of TCGA and Non-TCGA (ICGC). Although there are overlapping major histopathological subtypes such as ‘ductal’ and ‘lobular’ carcinoma, there are histopathological subtypes that are not perfectly matched with each other. More details of BRCA’s histopathological differences can be found in Supplementary Fig. S7. Developing a more robust cancer prediction method for inhomogeneous platforms in multi-site or multi-modal data would be an interesting future research direction.
Conclusion
We introduced a novel cancer type classification method, CPEM which is based on mutation features and an ensemble of machine learning classifiers. We conducted an in-depth study to clarify how various mutation feature groups affect the classification accuracy. Extensive investigation of various feature selection and classification methods based on machine learning algorithms led to the development of an ensemble model that classified 31 cancer types from the TCGA database at an average accuracy of 84% and six common cancer types at an average accuracy of 94%. The method outperforms the state-of-the-art mutation-based cancer classification methods.
Future plans are to utilize advanced deep neural networks to improve the cancer classification accuracy. Another idea we are currently exploring is to group cancer types in a hierarchical manner to reduce the search space size. We are also planning to apply our model to liquid biopsy data, such as circulating tumor DNA, and cells for primary tumor site prediction. Current liquid biopsy techniques are offered as laboratory-developed tests. Our method will be useful for early diagnosis of cancer as these liquid biopsy techniques evolve24. More rigorous validations using real clinical datasets will also be done.
Methods
Overview of the Proposed Method
CPEM consists of several data processing stages followed by a machine learning-based cancer classification stage, as shown in Fig. 1. The first step is constructing input feature vectors for various different types of somatic alterations detected from cancer genomic data to train and test machine learning classifiers. Since the input feature vectors are large and sparse (i.e., mostly zero), the next step is to reduce the size of the input feature set. This feature selection step is important to reduce the training time of the classifiers and also to increase the prediction accuracy by removing redundant and irrelevant features acting as noise. We explored several widely used feature selection algorithms25 and empirically chose a method that worked best for our dataset. The last step is the classification of the cancer type using machine learning classifiers. We employed an ensemble approach by combining a random forest and a deep neural network to maximize the classification accuracy. Detailed descriptions of each stage are provided in the following sections.
Feature Construction
The genetic alterations (e.g., mutation spectra, signatures and somatic copy number alterations (SCNAs)) often have a unique pattern depending on the cancer type8,9,26. Hence, these characteristics are sequentially applied to the subsequent analysis. We collected somatic single nucleotide variants (SNVs) and short insertions and deletions (indels) data for 31 TCGA cancer types from the GDAC27 website (Jan 28, 2016). We generated a data matrix of 22,421 genes × 7,115 samples that contains the number of protein sequencing altering SNVs and indels for each gene and sample. We also downloaded SCNA data from the GDAC website and constructed gene-level log2 copy ratio values to use them as additional input features. Mutation rates and mutation spectra/signatures were calculated using SNV data to be used as input features. As a result, we generated a data matrix of 35,565 input features × 7,002 samples to develop cancer type classification machine learning models. A pictorial description of our feature matrix is shown in Fig. 1, and the details of the input feature contents are as follows:
22,421 gene-level somatic mutation profile
2 mutation rates
6 mutation spectra (C > A, C > G, C > T, T > A, T > C, and T > G)
96 mutation signatures ({A, C, G, T} × 6 mutation spectra × {A, C, G, T})
13,040 gene-level log2 copy ratio
We created an initial prediction model using six machine learning models to evaluate the performance of the generated features. The initial results showed the highest accuracy in the random forest model. Subsequently, each feature was added to predict the accuracy of the initial model to verify the impact of each feature on the model.
Feature Selection
Feature selection is the process of reducing the dimension of the input feature data by removing redundant and irrelevant features that do not contribute to the final result. By selecting important features only, the classification can be more accurate and training time can be shortened. Various feature selection algorithms for genomic data have been proposed in the past25,28.
In this work, we tested several widely used supervised feature selection algorithms, including tree-based, least absolute shrinkage and selection operator (LASSO), and LSVC. We finally chose LSVC as the feature selection method (the various feature selection methods are compared above in ‘Comparison of Feature selection Methods’).
Tree-based feature selection
The tree-based feature selection method is based on the feature importance calculated during the training of the decision tree classifier, namely how much the feature contributes to the reduction of the overall error (e.g., accuracy or variance) of the classification. Once the importance of each feature is established, the features can be sorted and those with low importance can be discarded. In our experiment, we used the extremely randomized trees (extra trees)29, a decision forest that avoids the expensive bootstrapping process used in the random forest algorithm.
LASSO feature selection
LASSO is a linear regression problem with a L1-norm sparsity term defined as follow:
| 1 |
where xi is the training vector, yi is the predicted vector, and w is the regression coefficient vector.
After solving the minimization problem of Eq. 1, w serves as the importance value to filter the input feature xi. The number of selected features can be determined by the level of sparsity of w by changing the α parameter. If w is sparse then the small number of features will be selected, and vice versa.
LSVC feature selection
LSVC30 is based on a squared hinge loss function with an L1-norm sparsity term as follows:
| 2 |
| 3 |
where f is a linear decision function defined as fc(x) = wcTx + bc for each class c = 1,..., k in k-class. In our experiment, we choose the f as one-vs-rest decision function for multi-class classification. The loss function L is defined as follows:
| 4 |
Since w for each class will be sparse after minimization, important features can be selected by collecting elements with non-zero w. Similar to LASSO, the total number of selected features is determined by the sparsity of w, which can be controlled by changing ε.
Cancer Type Classification
Once feature data are collected and processed, the next step is training machine learning classifiers to predict cancer types. Most previous studies used a collection of binary classifiers (i.e., one-vs-rest)11,12,31 to predict the cancer types. Popular classifiers are random forests and support vector machine, with deep neural networks used most recently13. Rather than relying on one specific classifier, as in the previous studies, we proposed to build an ensemble of different machine learning methods, which is a widely used strategy to enhance the classification performance32,33. To achieve this, we tested four widely used machine learning classifiers, including random forests, OvR SVM, KNN, and fully connected DNN, and found the best classifiers for our ensemble model. To build an ensemble, we used multinomial classifiers rather than binary classifiers, and computed the average of the probability of each output to determine the per-label (i.e., cancer type) probability. We performed an in-depth analysis of the correlation of output from the classifiers (see ‘Putting it all together: CPEM results’ in the Results).
Supplementary information
Acknowledgements
We would like to thank Hahn Jo for proofreading the manuscript and Tran Minh Quan for validation of our implementation. This work is supported by the 2016 Research Fund (1.160052.01) and the 2019 Research Fund (1.190007.01 and 1.190033.01) of UNIST, the Next-Generation Information Computing Development Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (MSIT) (NRF-2016M3C4A7952635), the Collaborative Genome Program for Fostering New Post-Genome Industry of the NRF funded by the MSIT (NRF-2017M3C9A5031004), the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI) funded by the Ministry of Health & Welfare (HI18C0316), and the Basic Science Research Programs through the NRF funded by the MSIT (NRF-2018R1C1B6008448) and the Ministry of Education (NRF-2018R1A6A1A03025810).
Author contributions
S.L. and W.-K.J. designed the study. H.-O.J. prepared the data and tested the efficacy of mutation features. K.L. developed the machine learning algorithms and conducted the experiments. All authors analyzed the results and wrote the manuscript.
Data availability
The datasets and code used in this study are available on https://github.com/leekanggeun/CPEM.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Kanggeun Lee and Hyoung-oh Jeong.
Contributor Information
Semin Lee, Email: seminlee@unist.ac.kr.
Won-Ki Jeong, Email: wkjeong@unist.ac.kr.
Supplementary information
is available for this paper at 10.1038/s41598-019-53034-3.
References
- 1.Hudson, T. J., Anderson, W., Aretz, A. & Barker, A. D. International network of cancer genome projects. Nat. 464, 993–8, http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=2902243{&}tool=pmcentrez{&}rendertype=abstract{%}5Cnhttp://www.nature.com/nature/journal/v464/n7291/abs/nature08987.html., 10.1038/nature08987, NIHMS150003 (2010). [DOI] [PMC free article] [PubMed]
- 2.McLendon R, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nat. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinstein John N, Collisson Eric A, Mills Gordon B, Shaw Kenna R Mills, Ozenberger Brad A, Ellrott Kyle, Shmulevich Ilya, Sander Chris, Stuart Joshua M. The Cancer Genome Atlas Pan-Cancer analysis project. Nature Genetics. 2013;45(10):1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.International Cancer Genome Consortium et al. International network of cancer genome projects. Nat. 464, 993–8, https://www.ncbi.nlm.nih.gov/pubmed/20393554., 10.1038/nature08987 (2010). [DOI] [PMC free article] [PubMed]
- 5.Lawrence Michael S., Stojanov Petar, Mermel Craig H., Robinson James T., Garraway Levi A., Golub Todd R., Meyerson Matthew, Gabriel Stacey B., Lander Eric S., Getz Gad. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505(7484):495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoadley Katherine A., Yau Christina, Wolf Denise M., Cherniack Andrew D., Tamborero David, Ng Sam, Leiserson Max D.M., Niu Beifang, McLellan Michael D., Uzunangelov Vladislav, Zhang Jiashan, Kandoth Cyriac, Akbani Rehan, Shen Hui, Omberg Larsson, Chu Andy, Margolin Adam A., van’t Veer Laura J., Lopez-Bigas Nuria, Laird Peter W., Raphael Benjamin J., Ding Li, Robertson A. Gordon, Byers Lauren A., Mills Gordon B., Weinstein John N., Van Waes Carter, Chen Zhong, Collisson Eric A., Benz Christopher C., Perou Charles M., Stuart Joshua M. Multiplatform Analysis of 12 Cancer Types Reveals Molecular Classification within and across Tissues of Origin. Cell. 2014;158(4):929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greenman Christopher, Stephens Philip, Smith Raffaella, Dalgliesh Gillian L., Hunter Christopher, Bignell Graham, Davies Helen, Teague Jon, Butler Adam, Stevens Claire, Edkins Sarah, O’Meara Sarah, Vastrik Imre, Schmidt Esther E., Avis Tim, Barthorpe Syd, Bhamra Gurpreet, Buck Gemma, Choudhury Bhudipa, Clements Jody, Cole Jennifer, Dicks Ed, Forbes Simon, Gray Kris, Halliday Kelly, Harrison Rachel, Hills Katy, Hinton Jon, Jenkinson Andy, Jones David, Menzies Andy, Mironenko Tatiana, Perry Janet, Raine Keiran, Richardson Dave, Shepherd Rebecca, Small Alexandra, Tofts Calli, Varian Jennifer, Webb Tony, West Sofie, Widaa Sara, Yates Andy, Cahill Daniel P., Louis David N., Goldstraw Peter, Nicholson Andrew G., Brasseur Francis, Looijenga Leendert, Weber Barbara L., Chiew Yoke-Eng, deFazio Anna, Greaves Mel F., Green Anthony R., Campbell Peter, Birney Ewan, Easton Douglas F., Chenevix-Trench Georgia, Tan Min-Han, Khoo Sok Kean, Teh Bin Tean, Yuen Siu Tsan, Leung Suet Yi, Wooster Richard, Futreal P. Andrew, Stratton Michael R. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446(7132):153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alexandrov Ludmil B., Nik-Zainal Serena, Wedge David C., Aparicio Samuel A. J. R., Behjati Sam, Biankin Andrew V., Bignell Graham R., Bolli Niccolò, Borg Ake, Børresen-Dale Anne-Lise, Boyault Sandrine, Burkhardt Birgit, Butler Adam P., Caldas Carlos, Davies Helen R., Desmedt Christine, Eils Roland, Eyfjörd Jórunn Erla, Foekens John A., Greaves Mel, Hosoda Fumie, Hutter Barbara, Ilicic Tomislav, Imbeaud Sandrine, Imielinski Marcin, Jäger Natalie, Jones David T. W., Jones David, Knappskog Stian, Kool Marcel, Lakhani Sunil R., López-Otín Carlos, Martin Sancha, Munshi Nikhil C., Nakamura Hiromi, Northcott Paul A., Pajic Marina, Papaemmanuil Elli, Paradiso Angelo, Pearson John V., Puente Xose S., Raine Keiran, Ramakrishna Manasa, Richardson Andrea L., Richter Julia, Rosenstiel Philip, Schlesner Matthias, Schumacher Ton N., Span Paul N., Teague Jon W., Totoki Yasushi, Tutt Andrew N. J., Valdés-Mas Rafael, van Buuren Marit M., van ’t Veer Laura, Vincent-Salomon Anne, Waddell Nicola, Yates Lucy R., Zucman-Rossi Jessica, Andrew Futreal P., McDermott Ultan, Lichter Peter, Meyerson Matthew, Grimmond Sean M., Siebert Reiner, Campo Elías, Shibata Tatsuhiro, Pfister Stefan M., Campbell Peter J., Stratton Michael R. Signatures of mutational processes in human cancer. Nature. 2013;500(7463):415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lawrence Michael S., Stojanov Petar, Polak Paz, Kryukov Gregory V., Cibulskis Kristian, Sivachenko Andrey, Carter Scott L., Stewart Chip, Mermel Craig H., Roberts Steven A., Kiezun Adam, Hammerman Peter S., McKenna Aaron, Drier Yotam, Zou Lihua, Ramos Alex H., Pugh Trevor J., Stransky Nicolas, Helman Elena, Kim Jaegil, Sougnez Carrie, Ambrogio Lauren, Nickerson Elizabeth, Shefler Erica, Cortés Maria L., Auclair Daniel, Saksena Gordon, Voet Douglas, Noble Michael, DiCara Daniel, Lin Pei, Lichtenstein Lee, Heiman David I., Fennell Timothy, Imielinski Marcin, Hernandez Bryan, Hodis Eran, Baca Sylvan, Dulak Austin M., Lohr Jens, Landau Dan-Avi, Wu Catherine J., Melendez-Zajgla Jorge, Hidalgo-Miranda Alfredo, Koren Amnon, McCarroll Steven A., Mora Jaume, Lee Ryan S., Crompton Brian, Onofrio Robert, Parkin Melissa, Winckler Wendy, Ardlie Kristin, Gabriel Stacey B., Roberts Charles W. M., Biegel Jaclyn A., Stegmaier Kimberly, Bass Adam J., Garraway Levi A., Meyerson Matthew, Golub Todd R., Gordenin Dmitry A., Sunyaev Shamil, Lander Eric S., Getz Gad. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forbes SA, et al. COSMIC: Exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marquard AM, et al. TumorTracer: a method to identify the tissue of origin from the somatic mutations of a tumor specimen. BMC Med. Genomics. 2015;8:58. doi: 10.1186/s12920-015-0130-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Yukun, Sun Jingchun, Huang Liang-Chin, Xu Hua, Zhao Zhongming. Classification of Cancer Primary Sites Using Machine Learning and Somatic Mutations. BioMed Research International. 2015;2015:1–9. doi: 10.1155/2015/491502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan Y, et al. DeepGene: an advanced cancer type classifier based on deep learning and somatic point mutations. BMC Bioinforma. 2016;17:476. doi: 10.1186/s12859-016-1334-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kingma, D. P. & Ba, J. Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980 (2014).
- 15.Pedregosa F, et al. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 16.Kandoth Cyriac, McLellan Michael D., Vandin Fabio, Ye Kai, Niu Beifang, Lu Charles, Xie Mingchao, Zhang Qunyuan, McMichael Joshua F., Wyczalkowski Matthew A., Leiserson Mark D. M., Miller Christopher A., Welch John S., Walter Matthew J., Wendl Michael C., Ley Timothy J., Wilson Richard K., Raphael Benjamin J., Ding Li. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davies Helen, Bignell Graham R., Cox Charles, Stephens Philip, Edkins Sarah, Clegg Sheila, Teague Jon, Woffendin Hayley, Garnett Mathew J., Bottomley William, Davis Neil, Dicks Ed, Ewing Rebecca, Floyd Yvonne, Gray Kristian, Hall Sarah, Hawes Rachel, Hughes Jaime, Kosmidou Vivian, Menzies Andrew, Mould Catherine, Parker Adrian, Stevens Claire, Watt Stephen, Hooper Steven, Wilson Rebecca, Jayatilake Hiran, Gusterson Barry A., Cooper Colin, Shipley Janet, Hargrave Darren, Pritchard-Jones Katherine, Maitland Norman, Chenevix-Trench Georgia, Riggins Gregory J., Bigner Darell D., Palmieri Giuseppe, Cossu Antonio, Flanagan Adrienne, Nicholson Andrew, Ho Judy W. C., Leung Suet Y., Yuen Siu T., Weber Barbara L., Seigler Hilliard F., Darrow Timothy L., Paterson Hugh, Marais Richard, Marshall Christopher J., Wooster Richard, Stratton Michael R., Futreal P. Andrew. Mutations of the BRAF gene in human cancer. Nature. 2002;417(6892):949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 18.Kranenburg Onno. The KRAS oncogene: Past, present, and future. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 2005;1756(2):81–82. doi: 10.1016/j.bbcan.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 19.Hasanov E, Chen G, Chowdhury P, Weldon J, Ding Z, Jonasch E, Sen S, Walker C L, Dere R. Ubiquitination and regulation of AURKA identifies a hypoxia-independent E3 ligase activity of VHL. Oncogene. 2017;36(24):3450–3463. doi: 10.1038/onc.2016.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lokody Isabel. Reprogramming metabolic flux in glioma. Nature Reviews Cancer. 2014;14(11):706–707. doi: 10.1038/nrc3840. [DOI] [PubMed] [Google Scholar]
- 21.Turcan Sevin, Makarov Vladimir, Taranda Julian, Wang Yuxiang, Fabius Armida W. M., Wu Wei, Zheng Yupeng, El-Amine Nour, Haddock Sara, Nanjangud Gouri, LeKaye H. Carl, Brennan Cameron, Cross Justin, Huse Jason T., Kelleher Neil L., Osten Pavel, Thompson Craig B., Chan Timothy A. Mutant-IDH1-dependent chromatin state reprogramming, reversibility, and persistence. Nature Genetics. 2017;50(1):62–72. doi: 10.1038/s41588-017-0001-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holderfield Matthew, Deuker Marian M., McCormick Frank, McMahon Martin. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nature Reviews Cancer. 2014;14(7):455–467. doi: 10.1038/nrc3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fodde Riccardo, Smits Ron, Clevers Hans. APC, Signal transduction and genetic instability in colorectal cancer. Nature Reviews Cancer. 2001;1(1):55–67. doi: 10.1038/35094067. [DOI] [PubMed] [Google Scholar]
- 24.Cohen Joshua D., Li Lu, Wang Yuxuan, Thoburn Christopher, Afsari Bahman, Danilova Ludmila, Douville Christopher, Javed Ammar A., Wong Fay, Mattox Austin, Hruban Ralph H., Wolfgang Christopher L., Goggins Michael G., Dal Molin Marco, Wang Tian-Li, Roden Richard, Klein Alison P., Ptak Janine, Dobbyn Lisa, Schaefer Joy, Silliman Natalie, Popoli Maria, Vogelstein Joshua T., Browne James D., Schoen Robert E., Brand Randall E., Tie Jeanne, Gibbs Peter, Wong Hui-Li, Mansfield Aaron S., Jen Jin, Hanash Samir M., Falconi Massimo, Allen Peter J., Zhou Shibin, Bettegowda Chetan, Diaz Luis A., Tomasetti Cristian, Kinzler Kenneth W., Vogelstein Bert, Lennon Anne Marie, Papadopoulos Nickolas. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359(6378):926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hira Zena M., Gillies Duncan F. A Review of Feature Selection and Feature Extraction Methods Applied on Microarray Data. Advances in Bioinformatics. 2015;2015:1–13. doi: 10.1155/2015/198363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zack Travis I, Schumacher Steven E, Carter Scott L, Cherniack Andrew D, Saksena Gordon, Tabak Barbara, Lawrence Michael S, Zhang Cheng-Zhong, Wala Jeremiah, Mermel Craig H, Sougnez Carrie, Gabriel Stacey B, Hernandez Bryan, Shen Hui, Laird Peter W, Getz Gad, Meyerson Matthew, Beroukhim Rameen. Pan-cancer patterns of somatic copy number alteration. Nature Genetics. 2013;45(10):1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.The Broad Institute Genomic Data Analysis Centre., https://gdac.broadinstitute.org, https://gdac.broadinstitute.org (2016).
- 28.Huang L, Zhang HH, Zeng ZB, Bushel PR. Improved sparse multi-class SVM and its application for gene selection in cancer classification. Cancer Informatics. 2013;12:143–153. doi: 10.4137/CIN.S10212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Shen X. On l 1-norm multiclass support vector machines: methodology and theory. J. Am. Stat. Assoc. 2007;102:583–594. doi: 10.1198/016214506000001383. [DOI] [Google Scholar]
- 30.Geurts P, Ernst D, Wehenkel L. Extremely randomized trees. Mach. Learn. 2006;63:3–42. doi: 10.1007/s10994-006-6226-1. [DOI] [Google Scholar]
- 31.Kourou Konstantina, Exarchos Themis P., Exarchos Konstantinos P., Karamouzis Michalis V., Fotiadis Dimitrios I. Machine learning applications in cancer prognosis and prediction. Computational and Structural Biotechnology Journal. 2015;13:8–17. doi: 10.1016/j.csbj.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Competitions, K. https://www.kaggle.com/competitions (2017).
- 33.Dietterich TG, et al. Ensemble methods in machine learning. Multiple classifier systems. 2000;1857:1–15. doi: 10.1007/3-540-45014-9_1. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets and code used in this study are available on https://github.com/leekanggeun/CPEM.